Abstract
Introduction: Essential thrombocythemia (ET) is a classic BCL-ABL1-negative chronic myeloproliferative neoplasm (MPN), and is associated with an increased risk of hemorrhagic and thrombotic complications and leukemic transformation. Recently, two research groups discovered a high frequency of somatic calreticulin (CALR) mutations in patients with JAK2 V617F/MPL-unmutated ET and primary myelofibrosis. The pattern of most CALR mutations in MPNs is indels in exon 9 causing one base pair reading frameshift. CALR mutations have been found to have important diagnostic and prognostic significances in patients with ET. High-resolution melting analysis (HRMA) is a well-established method for the detection of or screening for mutations. The aims of this study were to screen for CALR mutations and to determine its clinical correlation in a cohort of adult Taiwanese patients with ET, and to test for the feasibility of HRMA as a screening method for the detection of CALR exon 9 mutations.
Methods: The screening for mutations in patients with hematologic neoplasms was approved by the Institutional Review Board of Mackay Memorial Hospital. 92 adult Taiwanese patients with ET were enrolled. Diagnosis of ET was established based on the 2008 WHO criteria. The clinical and laboratory characteristics of all patients at the time of diagnosis or referral were determined retrospectively by chart review. Genomic DNA derived from bone marrow, peripheral whole blood, and peripheral blood granulocytes or peripheral blood mononuclear cells were used for the screening. JAK2 V617F mutational status was screened by allele-specific polymerase chain reaction. CALR exon 9 mutations and MPL exon 10 mutations were screened by nucleotide sequencing on an ABI 3730 sequencer. The correlation between CALR mutational status and clinical characteristics was determined by using SPSS Statistics software (IBM, New York, USA). HRMA was performed with a CFX96 real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA).
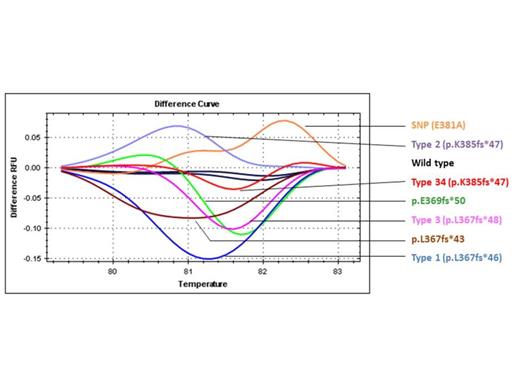
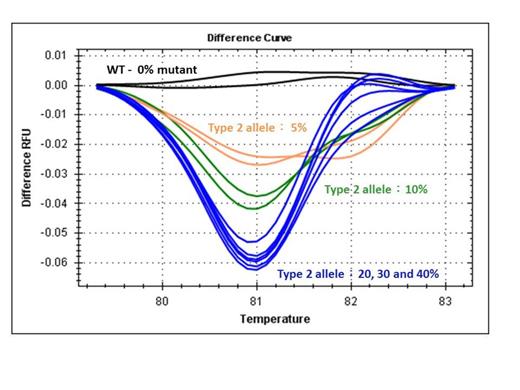
Results: In this cohort of ET patients, 59 (64%) patients harbored the JAK2 V617F mutation and one (1%) patient harbored the MPL W515K mutation. By Sanger sequencing, 17 (18% overall and 50% in JAK2/MPL-unmutated cases) patients harbored 6 types of CALR exon 9 mutations: 5 type 1 (p.L367fs*46), 6 type 2 (p.K385fs*47), 2 type 3 (p.L367fs*48), 2 type 34 (p.K385fs*47), and 2 novel types (p.L367fs*43 and p.E369fs*50). One patient with JAK2 V617F mutation was found to harbor a single nucleotide polymorphism in CALR exon 9 (c.1142 A>C, rs143880510). One patient with JAK2 V617F mutation also harbored a CALR type 3 mutation (p.L367fs*48), and was excluded from the correlation study. When compared with JAK2 V617F mutated ET patients, the presence of CALR mutations correlated with higher platelet count (median 1379 x10^9/L vs. 880 x10^9/L; P<0.001), lower hemoglobin level (median 12.5 g/dL vs. 13.6 g/dL; P=0.032), and younger age at diagnosis (median 44.5-year-old vs. 55-year-old; P=0.025). There was no significant difference between CALR mutated and JAK2 V617F mutated ET patients with regard to the frequency of thrombotic and hemorrhagic complications. The HRM curves of these CALR mutated patients could be distinguished from wild-type curves (Figure 1). Interestingly, the HRM curves of another 5 patients were also suspected for the presence of CALR exon 9 type 2 mutations but their results of CALR exon 9 Sanger sequencing were all wild-type. These 5 patients were suspected to have low CALR mutant allele burden. The sensitivity of HRMA to detect CALR exon 9 type 2 mutations was evaluated by serial dilution of a sample with approximately 40% mutant allele burden. CALR type 2 mutant could be detected in up to 5% dilution (Figure 2).
Conclusions: The frequency of CALR mutations and its phenotypic correlation in adult Taiwanese patients with ET is comparable with Western population. HRMA may be used as a rapid and relatively sensitive method for the screening of CALR mutations in patients with suspected MPN.
Representative melting curves of six CALR exon 9 mutations, one single nucleotide polymorphism (SNP) and wild-type DNA.
Representative melting curves of six CALR exon 9 mutations, one single nucleotide polymorphism (SNP) and wild-type DNA.
HRM curves of serial dilution of a CALR exon 9 type 2 mutant in a background of wild-type DNA.
HRM curves of serial dilution of a CALR exon 9 type 2 mutant in a background of wild-type DNA.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal