Abstract
Background: Cytomegalovirus (CMV) infection is one of the most common complications after hematopoietic cell transplantation (HCT). Regardless of preemptive therapy for CMV infection, CMV disease still occurs in approximately 10% of seropositive HCT recipients, resulting in high mortality. A number of studies have reported the association between CMV infection or disease and single-nucleotide polymorphisms (SNPs), however the results have not been validated in independent cohorts. We performed a candidate gene validation study of previously reported SNPs using a large genome-wide association study (GWAS) cohort.
Patients and methods: This GWAS cohort included donor-recipient (D/R) pairs who received the first allogeneic transplantation between 1990 and 2011 at the FHCRC. Of an overall cohort of 4855 D/R pairs, 2193 Caucasian CMV seropositive recipients and their donors (seropositve or seronegative) were analyzed. Two CMV phenotypes were analyzed: any infection within 100 days (only among 1585 recipients transplanted after 1995, antigenemia-based preemptive therapy administered) and CMV disease within 1 year after HCT. Published SNPs associated with CMV infection or disease were collected by a PubMed search using the keywords cytomegalovirus' and SNP' or polymorphism.' A p-value of less than 0.05 was required for validation. Genomic DNA samples from donors and recipients were analyzed using the Affymetrix GeneChip Genome-Wide Human SNP Array 5.0 (for one third of subjects) and Ilummina OmniExpress Beadchip Array (for the remainder). When the candidate SNPs were not included in the arrays, we imputed genotypes using IMPUTE software. Among genotyped and imputed SNPs, only SNPs with a minor allele frequency ≥0.05, call rate ≥90%, and Hardy-Weinberg equilibrium ≥0.0001 were analyzed. Donor serostatus, age, year of transplantation, donor type, HLA disparity, and intensity of conditioning regimen were used as adjustment factors as appropriate.
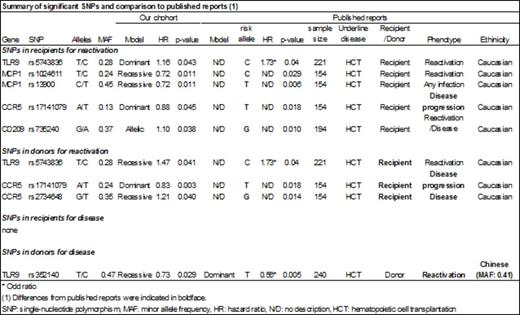
Results: Among the 2193 HCT recipients in our cohort, 1009/1585 (64%) had CMV infection and 354/2193 (16%) had CMV disease. We identified 52 SNPs in 31 publications that reported significant associations between SNPs and CMV infection or disease (median number of subjects in these studies, 215; range, 37-1770). A total of 32 SNPs were available for the analysis. For CMV infection, six SNPs in four genes (TLR9, MCP1, CCR5, and CD209) in recipient or donor met our validation criteria (Table). For all SNPs but one, however, the direction of the association was opposite to that reported in the previous studies. We validated one SNP association (TLR9/rs5743836) in the recipient exactly as originally reported (adjusted HR, 1.16; 95% CI, 1.00-1.34, p=0.043). The donor genotype at this SNP was also significantly associated with CMV infection (adjusted HR, 1.47; 95% CI, 1.02-2.14, p=0.041) (Table), but an association with donor genotype was not assessed in the original publication. As for CMV disease, a different TLR9 SNP (rs352140) in donors was identified as significant (adjusted HR, 0.74; 95% CI, 1.02-2.14, p=0.029), although the ethnicity is different from the reported cohort (Table). None of the candidate SNPs in the recipient genome were associated with CMV disease.
Conclusions: Of 32 SNPs that were previously reported to be associated with either CMV infection or disease, we were only able to validate two, both in TLR9. Most of the previously reported SNP associations for CMV infection or disease were not replicated in this large GWAS cohort. The inability to replicate these SNP associations does not necessarily negate the original discoveries, since many factors, including the sample size, could account for the different results. Our findings suggest that results of SNP association studies should be confirmed in multiple independent large cohorts and the biological mechanisms underlying the associations should be examined before clinical application. Unbiased discovery approaches are needed to determine the genetic associations of CMV infection and disease.
Boeckh:Chimerix Inc.: Consultancy, Research Funding; Viropharma Inc.: Research Funding; Genentech/Roche: Consultancy, Research Funding; Astellas: Consultancy, Research Funding; Merck: Consultancy, Research Funding; Gilead Sciences: Consultancy, Research Funding; Clinigen: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.
This icon denotes a clinically relevant abstract

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal