Abstract
Introduction: BCR-ABL mutations are the major background players in manifestation of resistance to all FDA approved tyrosine kinase inhibitors (TKIs) including imatinib, dasatinib, nilotinib, Bosutinib and ponatinib 1. Detection of mutations is a vital part of European or North American clinical guidelines at the time of resistance and/or drug switching, because resistance-causing mutations appearing as a result of one drug are sensitive to other in many instances and mutational data can therefore help in prescription of better alternative TKI in case of resistance or unsatisfactory response 1,2. Although BCR-ABL sequencing protocols are reported, they either lack the experimental details or are not cost-effective to be used in third world countries 1’3. Therefore, objective of this study was to develop a cost-effective protocol for BCR-ABL mutation detection in TKI resistant CML.
Material and Methods: Peripheral blood samples were collected from 10 imatinib resistant, 10 imatinib sensitive, 5 CML patients receiving nilotinib, positive for Philadelphia chromosome by conventional cytogenetics, and 10 healthy volunteers. Isolation of RNA was performed using TriZol® LS reagent and complementary DNA (cDNA) was prepared using random hexamer primers. The integrity of cDNA was checked by amplification of housekeeping gene GAPDH. A nested RT-PCR assay was optimized for ABL kinase domain amplification using standard PCR optimization techniques. PCR bands of 1306 or 1380 base pairs, corresponding to b2a2 and b3a2 BCR-ABL splice variants, were detected in 25 CML patients but no healthy controls. Consumables for CDNA and PCR were used from Fermentas (USA). PCR products were purified using Quick gel extraction kit (Invitrogen). DNA sequencing was performed using BigDye® Terminator v3.1Cycle Sequencing Kit (Applied Biosystems).
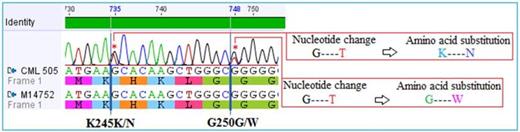
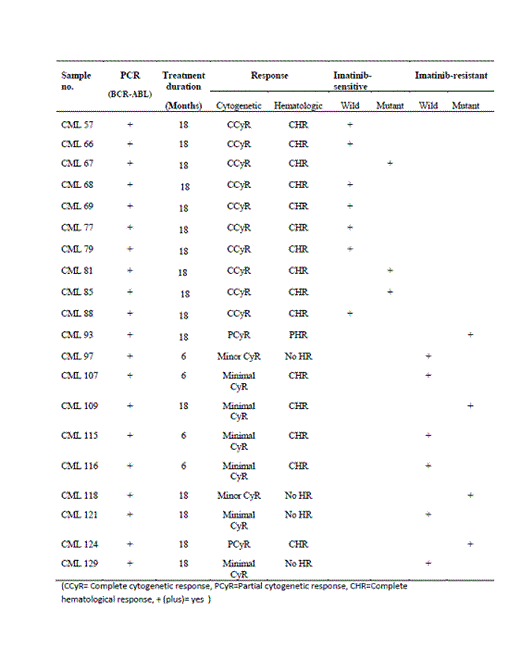
Results: Compound mutations were detected in CML patient showing primary resistance to nilotinib, including a novel K245N mutation and G250W mutations (Figure 1) while 4 of nilotinib responders did not show any mutations. Similarly, mutations detected in four (4/10, 40%) imatinib resistant were (G250W), (T394A), (Y253H), (E355G, Y393H}. Of 10 imatinib sensitive patients, mutations were detected in 3, 2 in accelerated phase and 1 in blast crisis, while none in 7 c chronic phase CML (Figure 2).
Discussion: We show association of BCR-ABL mutations with imatinib/nilotinib resistance and disease progression in CML patients, which is in accordance with previous studies 1’2’4,5. This also proves the usefulness and applicability of our BCR-ABL sequencing protocol for detection of clinically relevant mutations in CML patients receiving TKI treatment. A cost effective protocol it will facilitate the incorporation of mutation detection in clinical setting in low-resourced laboratories from third world countries and thus help better manage clinical interventions in drug-resistant CML 6’7.
References:
1. Baccarani M, Soverini S, De Benedittis C. Am Soc Clin Oncol Educ Book. 2014:167-75.
2. Kang Y, Hodges A, Ong E, Roberts W, Piermarocchi C, Paternostro G.PLoS One. 2014 Jul 16;9(7):e102221.
3. Branford S, Hughes T. Methods Mol Med. 2006; 125:93-106.
4. Viganò I, Di Giacomo N, Bozzani S, Antolini L, Piazza R, Gambacorti Passerini C. Am J Hematol. 2014 Jul 15.
5. Balabanov S, Braig M, Brümmendorf TH. Drug Discov Today Technol. 2014 Mar;11:89-99.
6. Jabbour E, Kantarjian H. Am J Hematol. 2014 May;89(5):547-56.
7. Kagita S, Jiwtani S, Uppalapati S, Linga VG, Gundeti S, Digumarti R. Tumour Biol. 2014 May;35(5):4443-6.
Electropherogram showing compound mutations, including a novel BCR-ABL mutation associated with primary nilotinib resistance in CML patient
Electropherogram showing compound mutations, including a novel BCR-ABL mutation associated with primary nilotinib resistance in CML patient
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal