Abstract
Introduction: Blood types result from genetic variation at blood group genes which directs the expression of blood group antigens on red blood cells. Determination of blood types is clinically important, as blood type mismatches can instigate potentially life-threatening allo-immune responses in transfusion recipients, transplant patients, and pregnant women. Blood types present in Asian American and Native American populations but rare or absent in the blood donor inventory, which is largely European American, can pose significant challenges to effective transfusion and pre-transfusion testing. We sought to more deeply characterize the blood type diversity present in Asian and Native Americans.
Methods: Consenting blood donors self-identified to be of Asian or Native American descent were eligible. Conventional serologic methods were used determine C, Jka, Jkb, M, and N blood types. Genotyping was performed with a blood type SNP array (HEA Beadchip, BioArray Solutions) to genetically assign c, C, e, E, K, k, Kpa, Kpb, Jsa, Jsb, Jka, Jkb, Fya, Fyb, , M, N, S, s, Lua, Lub, Dia, Dib, Coa, Cob, Doa, Dob, Joa, Hy, LWa, LWb, Sc1, and Sc2. For both methods, blood type was designated as positive + (consistent with presence of a blood group antigen) or negative 0 (consistent with absence of a blood group antigen). A SNP-serology discrepancy was defined as a + assignment by one method and a 0 by the other. Ambiguous SNP determinations were called + or 0 based upon relative signal intensities using established algorithms whenever possible; if not possible, the SNP-determined blood types were designated either inconclusive (IC) or low signal (LS).
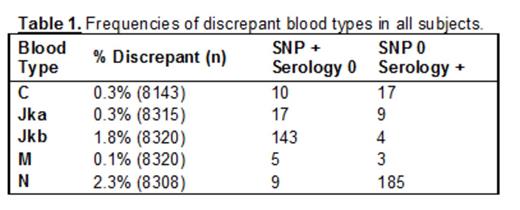
Results: A total of 8454 Asian and Native American blood donors representing 9 distinct ethnic groups were included in the study. As expected, the frequencies of rare and uncommon blood types differed between ethnic groups and in comparison to Europeans. Overall, 4.7% of blood donors studied exhibited one or more SNP-serology discrepancies for the four blood types tested by both methods (Table 1). The frequency of discrepancies was widely variable between blood types and between study populations (Figure 1), reaching as high as 5.4% for N (in Southeast Asians) and 7.2% for Jkb (in Pacific Islander/Hawaiians). We additionally observed patterns in IC and LS calls which varied between blood types and between ethnicities, suggesting that underlying genetic variation may contribute to inconclusive or low signal SNP results.
Conclusions: We characterized 8454 Asian and Native American blood donors for blood type by serology and with SNPs. As expected, we observed variation in the frequencies of blood type SNPs both between study populations and in comparison to Europeans. With additional testing, we found that 4.7% of donors exhibited discrepancies between SNP-predicted and serology-detected blood type, and that the frequencies of discrepancies varied between ethnic groups. We hypothesize that clinically relevant blood group gene variants were not accurately predicted using this SNP approach due to underlying genetic diversity at blood group loci in these populations. We propose that a more comprehensive approach, such as DNA sequencing, would characterize blood group gene variants in individuals of Asian and Native American heritage, as well as other genetically diverse populations.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal