Key Points
Increasing eIF2α phosphorylation increases fetal hemoglobin in human primary erythroid progenitors via a post-transcriptional mechanism.
Combining pharmacologic agents that use transcriptional and post-transcriptional mechanisms additively induces fetal hemoglobin.
Abstract
Strategies to increase fetal hemoglobin (HbF) levels can ameliorate symptoms and improve the lives of β-hemoglobinopathy patients. Although most studies have focused on induction of γ-globin gene expression as an approach to induce HbF, we hypothesized that post-transcriptional regulation of HbF plays an underappreciated yet important role in controlling HbF levels. In the present study, we investigated whether increasing eukaryotic initiation factor 2α (eIF2α) phosphorylation, a key regulator of protein translation, could enhance HbF post-transcriptionally in human primary erythroid cells. Initial analysis using a known inhibitor of eIF2α dephosphorylation, salubrinal, revealed that elevated eIF2α phosphorylation enhanced HbF production without changing globin gene expression, proliferation, or cell differentiation. These results were further supported by the post-transcriptional induction of HbF by other pharmacologic activators of the eIF2α pathway and by genetic inactivation of the negative regulators, GADD34 and CReP. Additionally, we found that this novel mechanism of increasing HbF could be combined with clinically relevant transcriptional activators of γ-globin gene expression to additively enhance HbF. Taken together, these findings identify eIF2α phosphorylation as a post-transcriptional regulator of HbF induction that may be pharmacologically targeted, either alone or in combination, in β-hemoglobinopathy patients.
Introduction
β-hemoglobinopathies, including sickle cell disease (SCD) and β-thalassemia (β-thal), are inherited disorders caused by mutations in the β-globin gene. These conditions result in dysfunctional or decreased β-globin protein, thereby causing severe anemia, organ damage, and reduced life expectancy.1-4 A promising therapeutic option with proven efficacy is the pharmacologic induction of fetal hemoglobin (HbF). This strategy developed from the observation that β-hemoglobinopathy patients with high levels of HbF have milder clinical disorders.5,6 This revealed γ-globin to be a suitable replacement for mutated or absent β-globin and proved that reactivation of the γ-globin gene is a viable therapeutic strategy.
There are more than 70 pharmacologic agents that induce γ-globin gene expression in a variety of model systems,7 and clinical trials have shown hydroxyurea (HU),8 butyrate,9,10 and DNA methyltransferase inhibitors11,12 to be effective pharmacologic inducers of HbF in β-hemoglobinopathy patients. However, no agent has the ideal combination of efficacy, safety, and ease of use.13,14 An interesting commonality of these compounds is that they all increase γ-globin through transcriptional mechanisms. Recently, there have been great advances in the understanding of the complex transcriptional program regulating hemoglobin switching with an underlying goal of discovering new mechanism-based therapeutic approaches to gene activation.15 In contrast, there is a small collection of data to suggest that post-transcriptional control may also be an important factor in hemoglobin production. For example, it has been shown that the stability of γ-globin messenger RNA (mRNA) is inversely related to the amount of β-globin mRNA16 and that enhanced γ-globin transcription does not always correlate with levels of γ-globin mRNA or HbF.17 Moreover, butyrate has been shown to increase the translational efficiency of γ-globin mRNA18 and 5-azacytidine (AZA) induces HbF to a greater degree than γ-globin mRNA steady state levels.19 These results suggest that post-transcriptional regulation of HbF plays an important but underappreciated role. A better understanding of this level of regulation could lead to new therapeutic targets and pharmacologic strategies that function through mechanisms that have never been previously used.
Because most HbF inducers are cytotoxic, we previously proposed that activation of cell stress signaling pathways has a central role in HbF induction.7 Based on this hypothesis, we chose to investigate whether the integrated stress response (ISR) pathway differentially regulates fetal and adult hemoglobin production post-transcriptionally. ISR signaling is centered on the eukaryotic initiation factor 2 (eIF2) and modulates translation initiation.20 In the presence of different cellular stresses, upstream eIF2α kinases are activated and phosphorylate eIF2 on the α-subunit at Ser51. Phosphorylated eIF2α (p-eIF2α) reduces global protein translation by inhibiting translation initiation but still allows translation of select transcripts that coordinate a stress response. In erythroid cells, the ISR pathway has been shown to modulate globin protein synthesis in response to heme availability and other stresses through the heme-regulated eIF2α kinase (HRI).21-23 Although not fully characterized in human erythroid cells, the HRI-eIF2α axis has been well-studied in murine models. Knockout experiments have demonstrated an important role of this pathway in regulating the severity of red blood cell disorders, such as iron deficiency anemia and β-thal.24,25 In the current study, we investigate the effects of ISR pathway activation in human erythroid progenitors as a potential mechanism of post-transcriptional induction of HbF.
Methods
Cell culture and chemicals
K562 cells were maintained in RPMI 1640 medium (Cellgro, Manassas, VA) supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin at 37°C with 5% CO2. granulocyte-colony stimulating factor-primed peripheral CD34+ cells were obtained from the hematopoietic cell processing core at the University of Washington or from Dr Patrick Gallagher of Yale Medical School using protocols approved by the institutional review board. Informed consent was obtained in accordance with the Declaration of Helsinki. Cultures were maintained as described in Sankaran et al.26 Salubrinal (Sal-003; Tocris Biosciences, Minneapolis, MN), guanabenz (GBZ) (Sigma-Aldrich, St. Louis, MO), HU (Sigma-Aldrich), and BTdCPU (kindly provided by Dr Bertal Aktas) were dissolved in dimethylsulfoxide and stored at −20°C. AZA (Sigma-Aldrich) was dissolved in phosphate-buffered saline and stored at −80°C.
Real-time quantitative reverse-transcription PCR and hemoglobin analysis
RNA was isolated from cells with RNeasy columns (Qiagen, Germantown, MD), cDNA was generated using the iScript complementary (c)DNA Synthesis Kit (Bio-Rad), and quantitative polymerase chain reaction (PCR) was performed with iQ SYBR Green Super Mix (Bio-Rad, Hercules, CA). Transcript levels were calculated by the method of Larionov et al27 relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression. PCR primers are listed in Table 1.
Quantitative real-time RT-PCR primers
| Gene . | Forward (5′ → 3′) . | Reverse (5′ → 3′) . |
|---|---|---|
| ATF3 | CTCGGAAGTGAGTGCTTCTG | CCGTCTTCTCCTTCTTCTTG |
| CHOP | GACCTGCAAGAGGTCCTGTC | TCGCCTCTACTTCCCTGGTC |
| CREP | CCGCATTCGCTGCCCTCTGT | CGTTTCCGCGATCCGCCTGT |
| GADD34 | CTGAGCCCTGCCCCTTCCGA | GAAGCGCACCTTTCTGGCCTT |
| GAPDH | TCCCATCACCATCTTCCA | CATCACGCCACAGTTTCC |
| β-globin | GGTGGTCTACCCTTGGACCC | GATACTTGTGGGCCAGGGCA |
| γ-globin | AGACGCCATGGGTCATTTCACA | GCCTATCCTTGAAAGCTCTGCAT |
| Gene . | Forward (5′ → 3′) . | Reverse (5′ → 3′) . |
|---|---|---|
| ATF3 | CTCGGAAGTGAGTGCTTCTG | CCGTCTTCTCCTTCTTCTTG |
| CHOP | GACCTGCAAGAGGTCCTGTC | TCGCCTCTACTTCCCTGGTC |
| CREP | CCGCATTCGCTGCCCTCTGT | CGTTTCCGCGATCCGCCTGT |
| GADD34 | CTGAGCCCTGCCCCTTCCGA | GAAGCGCACCTTTCTGGCCTT |
| GAPDH | TCCCATCACCATCTTCCA | CATCACGCCACAGTTTCC |
| β-globin | GGTGGTCTACCCTTGGACCC | GATACTTGTGGGCCAGGGCA |
| γ-globin | AGACGCCATGGGTCATTTCACA | GCCTATCCTTGAAAGCTCTGCAT |
Hemoglobin analysis was performed using 2 × 106 cells per sample. Hemoglobin content was calculated from the absorbance at 415 nm using the extinction coefficient of 125 mM-1cm−1.28 Hemoglobin high-performance liquid chromatography (HPLC) was performed as described in Ou and Rognerud29 using a PolyCAT-A cation exchange column (The Nest Group, Southborough, MA).
Flow cytometry and cell morphology
Cells were stained with CD71-APC (BD Biosciences 551374, Franklin Lakes, NJ) and CD235a-fluorescein isothiocyanate (FITC) (BD Biosciences 559943) and were analyzed by a BD FACSCalibur cytometer. Fluorescence gating was set by matched isotype controls fluorescein isothiocyanate (FITC) mouse IgG2b,κ (BD Biosciences 555742) and APC mouse IgG2a,κ (BD Biosciences 555576). To determine lentiviral infection efficiency, green fluorescent protein fluorescence was determined using a BD FACSCalibur cytometer for transduced samples and was compared with an untransduced control. All data were analyzed using FlowJo software (TreeStar).
Cell morphology was evaluated by staining with Modified Wright-Giemsa stain (Sigma) under light microscopy at 40× magnification using an Olympus BX51 microscope and Image-Pro Plus 7.0 software.
Western blot analysis
Cells were lysed in Cell Signaling Lysis Buffer (Cell Signaling, Danvers, MA) containing complete, Mini, EDTA-free Protease Inhibitor Tablet (Roche Diagnostics, Indianapolis, IN) and phosphatase inhibitors (Sigma-Aldrich P5726, P0044), resolved by electrophoresis, and transferred to Immobilon-P membranes (Millipore, Billerica, MA). Blots were incubated overnight at 4°C in the following antibodies: p-eIF2α (1:1000; Cell Signaling 3398), eIF2α (1:1000; Santa Cruz sc-11386), GAPDH (1:2000; Santa Cruz sc-47724), Activating Transcription Factor 4 (ATF4) (1:500; Santa Cruz sc-22800), Growth Arrest and DNA-Damaging Inducible Protein 34 (GADD34) (1:1500; Proteintech 10449-1-AP, Chicago, IL), Constitutive Repressor of eIF2α Phosphorylation (CReP) (1:1000; Proteintech 14634-1-AP), and β-actin (1:20 000; Sigma A1978).
Lentiviral vectors and infection
Lentiviral vectors were obtained from The RNAi Consortium (Broad Institute). Oligonucleotides were cloned into a pLKO_TRC019 vector containing a green fluorescent protein selection marker as previously described in Moffat et al.30 The sequence targeted by GADD34 short hairpin RNA (shRNA) was 5′-CGAGAAGGTCACTGTCCATTT-3′ and CReP shRNA was 5′-GAACCTGCATCCATCCCTTGCA-3′. The vector pLKO_TRC029 containing a nullT sequence31 instead of a gene targeting shRNA was used as a negative control. The virus was generated according to The RNAi Consortium protocols (Broad Institute). For infections on day 12 of the culture, the virus was concentrated by centrifugation at 100 000 × g at 4°C for 1 hour and the virus pellets were resuspended in phosphate-buffered saline. The cells were plated (1 × 106 cells/well) in 12-well plates and spin-infected for 30 minutes at 2300 rpm with concentrated virus and 8 μg/mL polybrene (Sigma-Aldrich). After overnight incubation, this process was repeated the following day with fresh virus, and cells were returned to normal culture medium 24 hours after the second infection.
Results
Salubrinal activates the ISR pathway in K562 cells
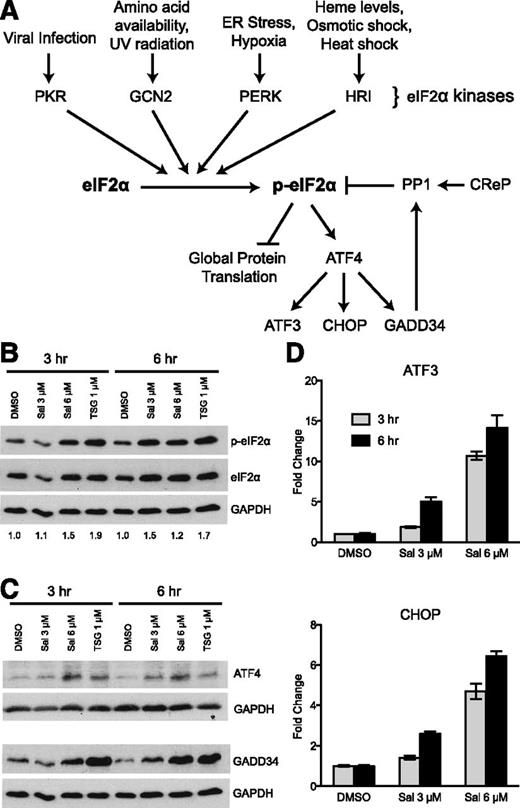
To begin testing our hypothesis, we used a known pharmacologic activator of the ISR pathway, salubrinal (Sal). We chose Sal because it has been previously shown to inhibit the important negative feedback loop that regulates dephosphorylation of p-eIF2α,32 thus increasing p-eIF2α without exposure to harmful cell stresses. Normally under stress conditions, eIF2α is phosphorylated and global translation is inhibited, but a few transcripts are selectively translated (Figure 1A). A well-characterized example is the selective translation of ATF4 via multiple upstream open reading frames located in its 5′-untranslated region.33,34 ATF4 is a transcription factor that upregulates many stress response genes, including ATF3, CHOP, and GADD34. GADD34 protein, acting as a chaperone, then binds to protein phosphatase 1 (PP1) and targets it to dephosphorylate p-eIF2α and decreases the stress response.35 Another chaperone, CReP, also targets PP1 to dephosphorylate p-eIF2α, but is constitutively present in the cell and does not require stress to be induced.36 Sal reduces the negative regulation of this pathway by inhibiting the ability of GADD34- and CReP-PP1 complexes to dephosphorylate p-eIF2α.
The ISR pathway is activated by salubrinal in K562 cells. (A) A simplified diagram of the ISR pathway showing both activation and negative regulatory components. (B) Western blot analysis reveals 3 μM and 6 μM salubrinal (Sal) enhances p-eIF2α at 3 and 6 hours. These results were compared with 1 μM TSG as a positive control. Numbers below each lane denote the ratio of p-eIF2α/eIF2α quantified by ImageJ software. GAPDH and total eIF2α were evaluated as loading controls. (C) 3 μM and 6 μM Sal augment downstream ISR signaling to a similar extent as 1 μM TSG, as shown by western blot analysis of ATF4 and GADD34 relative to GAPDH loading control. (D) Sal increases the mRNA expression of ATF3 and CHOP dose dependently at 3 and 6 hours post-treatment. Transcript levels at each time point are reported as fold increase relative to a dimethylsulfoxide (DMSO) control. Error bars denote ± standard deviation of 3 technical replicates.
The ISR pathway is activated by salubrinal in K562 cells. (A) A simplified diagram of the ISR pathway showing both activation and negative regulatory components. (B) Western blot analysis reveals 3 μM and 6 μM salubrinal (Sal) enhances p-eIF2α at 3 and 6 hours. These results were compared with 1 μM TSG as a positive control. Numbers below each lane denote the ratio of p-eIF2α/eIF2α quantified by ImageJ software. GAPDH and total eIF2α were evaluated as loading controls. (C) 3 μM and 6 μM Sal augment downstream ISR signaling to a similar extent as 1 μM TSG, as shown by western blot analysis of ATF4 and GADD34 relative to GAPDH loading control. (D) Sal increases the mRNA expression of ATF3 and CHOP dose dependently at 3 and 6 hours post-treatment. Transcript levels at each time point are reported as fold increase relative to a dimethylsulfoxide (DMSO) control. Error bars denote ± standard deviation of 3 technical replicates.
First we performed a dose response of Sal in the K562 erythroleukemia cell line to identify its lowest doses that enhanced p-eIF2α levels and downstream ISR signaling. Within 6 hours, 3 and 6 μM Sal increased p-eIF2α to a similar extent as our positive control, an endoplasmic reticulum stress-inducer thapsigargin (TSG) (Figure 1B). Correspondingly, Sal also enhanced ATF4 and GADD34 protein (Figure 1C), as well as mRNA expression of ATF3 and CHOP in a dose-dependent manner (Figure 1D). These results identified doses of Sal that are capable of increasing p-eIF2α to an extent that activates ISR signaling.
Salubrinal increases p-eIF2α and inhibits translation initiation in human primary erythroid progenitors
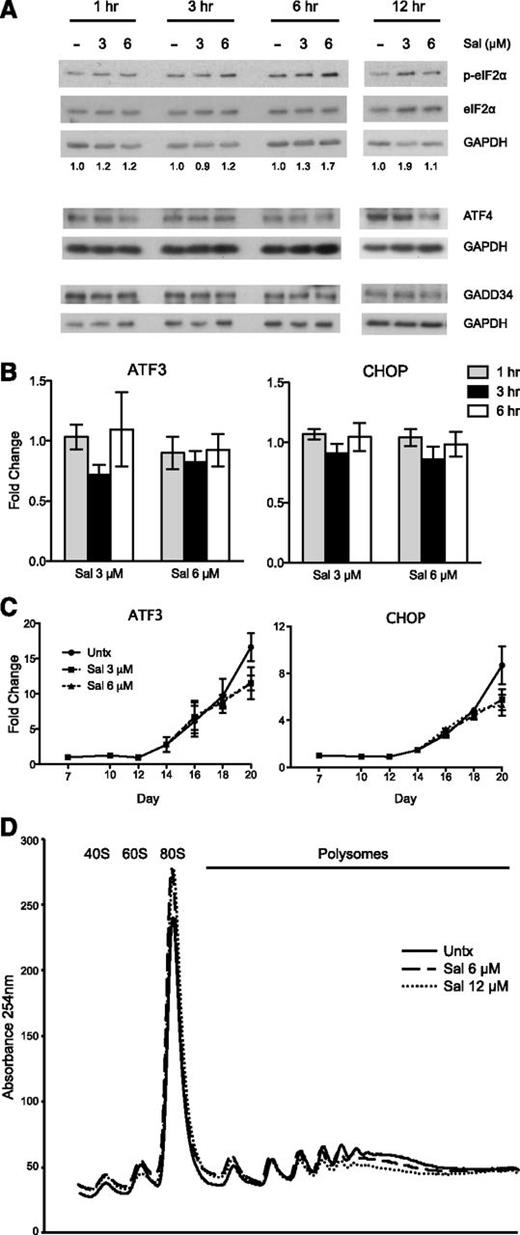
To be able to adequately test the ISR pathway in a physiologically relevant context containing both HbF and adult hemoglobin (HbA), we used a previously described 2-phase in vitro erythroid differentiation system.26 granulocyte-colony stimulating factor-mobilized CD34+ peripheral blood cells were expanded for a week in culture, and erythropoietin was added to the medium on day 7 to facilitate erythroid differentiation through day 20. Our first objective was to verify that 3 and 6 μM Sal could increase p-eIF2α in these cells. Indeed, Sal enhanced p-eIF2α levels within a 12-hour course of time when treated on day 15 of the culture (Figure 2A). Although this increase was not as dramatic as seen in K562 cells, it is important to note that phosphorylation of only 20% to 30% of eIF2α is needed to completely inhibit protein synthesis.37,38 Next, we analyzed downstream ISR signaling and were surprised to find that Sal did not increase ATF4 or GADD34 protein (Figure 2A). In fact, 6 μM Sal slightly reduced ATF4 at 12 hours after treatment compared with the untreated control. In line with these data, there was no increase in ATF3 or CHOP gene expression when analyzed within 6 hours (Figure 2B) or during the entire differentiation period (Figure 2C).
The effects of salubrinal on the ISR pathway and protein translation in human primary erythroid progenitors. (A) Western blot analysis reveals p-eIF2α is increased with salubrinal (Sal) treatment on day 15 of the culture. In contrast, ATF4 and GADD34 protein are not increased with Sal treatment relative to an untreated control (-). Numbers below each lane denote the ratio of p-eIF2α/eIF2α quantified by ImageJ software. Total eIF2α and GAPDH are used as loading controls. (B) The mRNA expression of ATF3 and CHOP are not increased at 1 to 6 hours after Sal treatment on day 15. Expression is reported as fold change relative to untreated negative control. Error bars represent ± standard error of the mean of 3 independent experiments (2 unique donors). (C) ATF3 and CHOP mRNA levels are not enhanced with Sal treatment over the course of differentiation. Expression is reported as fold change relative to the untreated control (Untx) on day 7. Sal (3 μM and 6 μM) were treated on days 15 and 18. Error bars represent ± standard error of the mean of 3 independent experiments (2 different donors). (D) Representative polysome profile on day 18 after 6 hours of Sal treatment reveals a polysome to monosome shift, indicative of halted translation initiation and reduced translation.
The effects of salubrinal on the ISR pathway and protein translation in human primary erythroid progenitors. (A) Western blot analysis reveals p-eIF2α is increased with salubrinal (Sal) treatment on day 15 of the culture. In contrast, ATF4 and GADD34 protein are not increased with Sal treatment relative to an untreated control (-). Numbers below each lane denote the ratio of p-eIF2α/eIF2α quantified by ImageJ software. Total eIF2α and GAPDH are used as loading controls. (B) The mRNA expression of ATF3 and CHOP are not increased at 1 to 6 hours after Sal treatment on day 15. Expression is reported as fold change relative to untreated negative control. Error bars represent ± standard error of the mean of 3 independent experiments (2 unique donors). (C) ATF3 and CHOP mRNA levels are not enhanced with Sal treatment over the course of differentiation. Expression is reported as fold change relative to the untreated control (Untx) on day 7. Sal (3 μM and 6 μM) were treated on days 15 and 18. Error bars represent ± standard error of the mean of 3 independent experiments (2 different donors). (D) Representative polysome profile on day 18 after 6 hours of Sal treatment reveals a polysome to monosome shift, indicative of halted translation initiation and reduced translation.
Because Sal failed to activate canonical downstream ISR gene expression, we used polysome profiling to determine if the increased p-eIF2α was affecting translation. This technique can determine if protein translation is decreased, as indicated by a shift in mRNA from the polysome to monosome fractions when translation initiation is inhibited.39 When primary erythroid progenitors were treated with Sal for 6 hours, polysome-associated mRNA was reduced, coincident with an increased 80S monosome peak (Figure 2D). Taken together, these data indicate that Sal increases p-eIF2α to an extent that is sufficient to repress translation initiation.
Salubrinal increases HbF via a post-transcriptional mechanism
Next, we optimized a Sal dosing schedule to evaluate its effects on globin production. We found that Sal treatments on days 15 and 18 of culture were well tolerated and did not change the differentiation profile (Figure 3A-C) or alter cellular proliferation (Figure 3D). To confirm that these effects were not due to time of treatment, cells were also treated on days 9 and 12 and no differences were found in cell proliferation or differentiation (supplemental Figures 1A-B). This is an important point, as changes in growth and differentiation could alter HbF production, independent of changes in p-eIF2α.
Salubrinal does not change cellular differentiation, proliferation, or globin gene expression in human primary erythroid cells. (A) Fluorescence-activated cell sorter analysis was performed with antibodies recognizing CD71 and CD235a on day 20. This representative experiment shows that salubrinal (Sal) does not change the overall differentiation profile relative to the untreated control (Untx). (B) Flow cytometry was performed in five independent experiments using two different donors to quantify CD71 and CD235a cell surface markers on day 20. Salubrinal (Sal) (3 μM and 6 μM) were compared with the untreated control. This box-and-whisker plot shows the distribution of the percent of cells that are double positive for CD71 and CD235a. The lines inside the box show the median, the boxes show the quartiles, and the whiskers show the extremes of the observed percentages. (C) Modified Wright-Giemsa staining of untreated and Sal treated cells on day 19 reveals similar stages of erythroid differentiation. Images were obtained with an Olympus BX51 microscope, 40× magnification, and Image-Pro Plus 7.0 software. (D) Sal does not alter cell growth during the treatment period. Sal was treated on days 15 and 18 and the fold increase of cell growth was compared with the untreated control on day 15. Cells were counted on days 15, 17, and 20 using trypan blue exclusion. Error bars represent ± SEM of three independent experiments (two unique donors). (E) The mRNA expression of γ- and β-globin was evaluated over the course of differentiation from day 7 through day 20. Sal treatments (3 μM and 6 μM) occurred on days 15 and 18, and the fold increase relative to the untreated control day 7 sample is shown. No significant changes in γ-globin or β-globin mRNA levels are observed. Error bars represent ± standard error of the mean of 3 independent experiments (2 different donors). (F) The AUC was calculated for γ- and β-globin mRNA expression from days 7 through 20. This representation of the data allows quantification of the total mRNA from each gene available for translation over the course of erythroid differentiation. Sal treatments (3 μM and 6 μM) on days 15 and 18 did not change the γ- or β-globin AUC relative to the untreated control. Error bars represent ± standard error of the mean of 3 independent experiments (2 different donors). (G) The AUCs for γ-globin and β-globin were compared as a γ/(γ + β) ratio. Sal treatment (3 μM and 6 μM) did not change this ratio relative to the untreated control in 3 independent experiments using 2 unique donors. Error bars denote ± standard error of the mean.
Salubrinal does not change cellular differentiation, proliferation, or globin gene expression in human primary erythroid cells. (A) Fluorescence-activated cell sorter analysis was performed with antibodies recognizing CD71 and CD235a on day 20. This representative experiment shows that salubrinal (Sal) does not change the overall differentiation profile relative to the untreated control (Untx). (B) Flow cytometry was performed in five independent experiments using two different donors to quantify CD71 and CD235a cell surface markers on day 20. Salubrinal (Sal) (3 μM and 6 μM) were compared with the untreated control. This box-and-whisker plot shows the distribution of the percent of cells that are double positive for CD71 and CD235a. The lines inside the box show the median, the boxes show the quartiles, and the whiskers show the extremes of the observed percentages. (C) Modified Wright-Giemsa staining of untreated and Sal treated cells on day 19 reveals similar stages of erythroid differentiation. Images were obtained with an Olympus BX51 microscope, 40× magnification, and Image-Pro Plus 7.0 software. (D) Sal does not alter cell growth during the treatment period. Sal was treated on days 15 and 18 and the fold increase of cell growth was compared with the untreated control on day 15. Cells were counted on days 15, 17, and 20 using trypan blue exclusion. Error bars represent ± SEM of three independent experiments (two unique donors). (E) The mRNA expression of γ- and β-globin was evaluated over the course of differentiation from day 7 through day 20. Sal treatments (3 μM and 6 μM) occurred on days 15 and 18, and the fold increase relative to the untreated control day 7 sample is shown. No significant changes in γ-globin or β-globin mRNA levels are observed. Error bars represent ± standard error of the mean of 3 independent experiments (2 different donors). (F) The AUC was calculated for γ- and β-globin mRNA expression from days 7 through 20. This representation of the data allows quantification of the total mRNA from each gene available for translation over the course of erythroid differentiation. Sal treatments (3 μM and 6 μM) on days 15 and 18 did not change the γ- or β-globin AUC relative to the untreated control. Error bars represent ± standard error of the mean of 3 independent experiments (2 different donors). (G) The AUCs for γ-globin and β-globin were compared as a γ/(γ + β) ratio. Sal treatment (3 μM and 6 μM) did not change this ratio relative to the untreated control in 3 independent experiments using 2 unique donors. Error bars denote ± standard error of the mean.
Then we analyzed γ- and β-globin gene expression throughout differentiation. Treatment with either 3 or 6 μM Sal on days 15 and 18 did not significantly change γ- or β-globin mRNA expression relative to the untreated control (Figure 3E). We also compared the area under the curve (AUC) of the globin mRNA levels, which is calculated from days 7 to 20 for γ- and β-globin (Figure 3F) and compared as a γ/(γ + β) ratio (Figure 3G). The AUC estimates the total amount of mRNA for each gene that is available for translation throughout the differentiation period. We found that Sal treatment did not change the γ/(γ + β) ratio, confirming that Sal did not change globin mRNA expression. To ensure this result was not due to being treated late in erythropoiesis, cells were treated with Sal on days 9 and 12, which also did not change γ- or β-globin mRNA levels (supplemental Figure 1C-D).
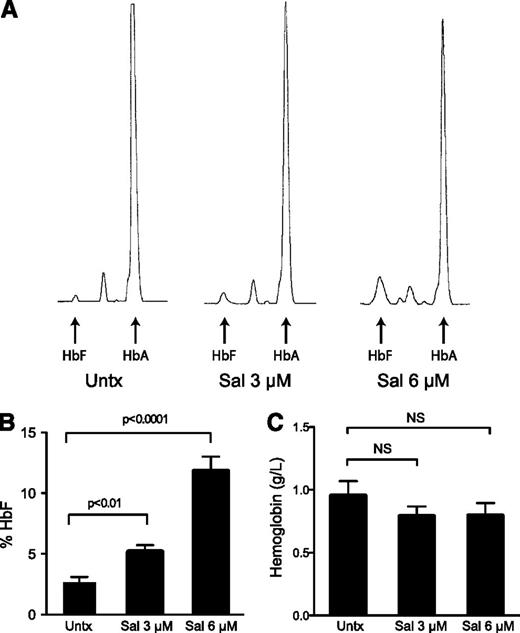
In contrast to the transcript expression, 3 and 6 μM Sal dose dependently increased the percentage of HbF (Figure 4A-B) and absolute levels of HbF (supplemental Figure 2A) when evaluated by HPLC at the end of the culture. Compared with untreated cells, 3 μM Sal increased the percent HbF from 2.6% to 5.1% (2-fold) and 6 μM Sal resulted in 11.8% HbF (4.5-fold). Considering the unchanged globin mRNA expression, these results suggest that Sal increases HbF by a post-transcriptional mechanism. Representative HPLC traces reveal that the enhanced percentages were due to increased HbF, but also reduced HbA, providing evidence that HbF and HbA may be differentially or reciprocally regulated at the translational level. Sal treatments earlier in the differentiation process, before considerable amounts of hemoglobin are synthesized, failed to significantly increase HbF, further supporting this conclusion (supplemental Figure 1E). Additionally, we wanted to ensure total hemoglobin levels were not dramatically decreased because we previously confirmed that Sal reduced global protein synthesis. We found that there was a trend for Sal treatment to slightly reduce total hemoglobin content by 16.8%, although these results were not significantly different from the untreated control across 5 independent experiments (Figure 4C).
Salubrinal induces HbF in human erythroid progenitors. (A) Differentiating erythroid progenitors were treated with 3 μM and 6 μM salubrinal (Sal) on days 15 and 18 of the culture. On day 20, cells were lysed and the proportions of HbA and HbF were determined by ion-exchange HPLC. These HPLC traces from a representative experiment reveal that Sal enhances HbF, as well as it reduces HbA compared with the untreated control. (B) The percentage of HbF was quantified in 5 independent experiments (3 different donors) comparing 3 μM and 6 μM Sal treatments to an untreated control. Sal dose-dependently increases the percentage of HbF. Error bars express ± standard error of the mean and P values were calculated using an unpaired two-tailed t test. (C) Equal numbers of cells were lysed on day 20 and hemoglobin concentrations were determined by measuring the absorbance at 415 nm. Sal treatments did not significantly reduce total hemoglobin content. Error bars represent ± standard error of the mean of 5 independent experiments (3 unique donors). P values were calculated using an unpaired two-tailed t test and were found to be nonsignificant (NS). The P value for 3 μM and 6 μM Sal compared with untreated was P = .26 and P = .32, respectively.
Salubrinal induces HbF in human erythroid progenitors. (A) Differentiating erythroid progenitors were treated with 3 μM and 6 μM salubrinal (Sal) on days 15 and 18 of the culture. On day 20, cells were lysed and the proportions of HbA and HbF were determined by ion-exchange HPLC. These HPLC traces from a representative experiment reveal that Sal enhances HbF, as well as it reduces HbA compared with the untreated control. (B) The percentage of HbF was quantified in 5 independent experiments (3 different donors) comparing 3 μM and 6 μM Sal treatments to an untreated control. Sal dose-dependently increases the percentage of HbF. Error bars express ± standard error of the mean and P values were calculated using an unpaired two-tailed t test. (C) Equal numbers of cells were lysed on day 20 and hemoglobin concentrations were determined by measuring the absorbance at 415 nm. Sal treatments did not significantly reduce total hemoglobin content. Error bars represent ± standard error of the mean of 5 independent experiments (3 unique donors). P values were calculated using an unpaired two-tailed t test and were found to be nonsignificant (NS). The P value for 3 μM and 6 μM Sal compared with untreated was P = .26 and P = .32, respectively.
Complementary pharmacologic and genetic approaches to ISR activation confirm involvement of p-eIF2α in post-transcriptional HbF induction mechanism
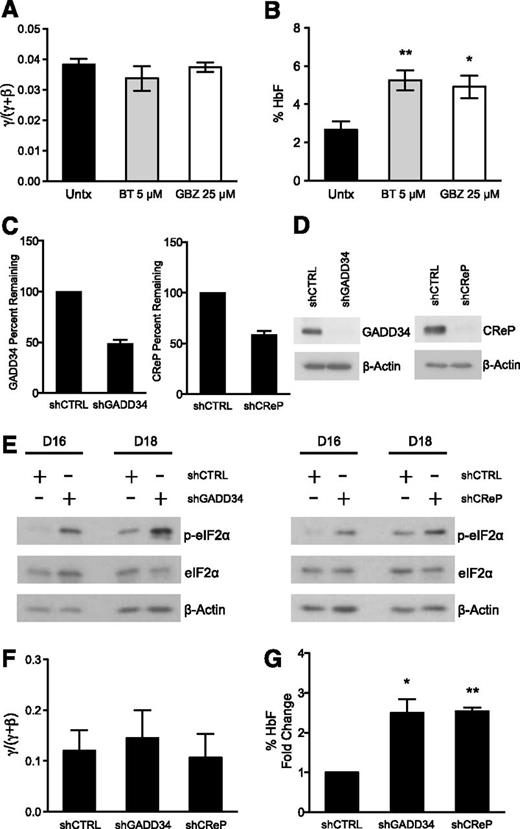
To further test whether increased p-eIF2α enhances HbF production post-transcriptionally, we assessed other pharmacologic agents that target the ISR pathway. We chose 2 compounds that have been shown to increase p-eIF2α through unique mechanisms. BTdCPU (BT) was discovered in a chemical genetics screen to increase p-eIF2α by directly activating the HRI kinase,40 the critical eIF2 kinase in erythroid cells. GBZ is an anti-hypertensive agent that was recently found to disrupt the association of GADD34 and PP1 to sustain p-eIF2α, attenuate protein translation, and restore protein homeostasis in stressed cells.41 We treated differentiating primary erythroid precursors with 5 μM BT and 25 μM GBZ on days 15 and 18. Similarly to Sal, BT and GBZ did not change the γ/(γ + β) ratio of globin mRNA expression (Figure 5A), but it did result in a 2-fold increase in the percentage of HbF on day 20 (Figure 5B). BT also increased the absolute levels of HbF such as Sal, but GBZ failed to do so, indicating that this drug primarily reduced HbA (supplemental Figure 2B).
Complementary pharmacologic and genetic methods augment p-eIF2α and increase HbF. (A) Human primary erythroid cells were treated with 5 μM BT and 25 μM GBZ on days 15 and 18 of differentiation. The mRNA expression for γ- and β-globin was assessed from day 7 through day 20 and the AUC was calculated for each. BT and GBZ treatments did not change the γ/(γ + β) AUC mRNA ratio relative to untreated control. The error bars represent ± standard error of the mean of 3 independent experiments assessing BT treatment and 2 independent experiments analyzing the effects of GBZ. (B) HPLC was used to determine the amount of HbA and HbF present on day 20 from lysates prepared from erythroid progenitors treated with 5 μM BT and 25 μM GBZ. Both BT and GBZ significantly increase the percentage of HbF relative to the untreated control. Error bars represent ± standard error of the mean of 3 independent experiments assessing BT treatment and 2 independent experiments analyzing GBZ-treated cells. P values were determined using an unpaired two-tailed t test. *P < .05; **P < .01. (C) Differentiating human primary erythroid progenitors were infected with a control shRNA (shCTRL) and shRNAs targeting either GADD34 (shGADD34) or CReP (shCReP) on days 12 and 13. On day 16, transcript levels were evaluated for GADD34 and CReP using quantitative PCR. Both shGADD34 and shCReP resulted in approximately 50% knockdown when normalized to the shCTRL. Error bars denote ± standard error of the mean of 3 independent experiments (3 different donors). (D) Western blot analysis shows shGADD34 and shCReP result in significant knockdown of GADD34 and CReP protein, respectively, in comparison with shCTRL. Protein lysates were taken on day 16 of differentiation after 2 infections on days 12 and 13. As a loading control, β-actin was used. (E) Protein lysates were taken on days 16 and 18 of differentiation from erythroid progenitors infected with shCTRL, shGADD34, or shCReP. Western blot analysis reveals both shGADD34 and shCReP significantly increase p-eIF2α levels compared with shCTRL. Total eIF2α and β-actin were used as loading controls. (F) The mRNA expression of γ- and β-globin was assessed throughout differentiation. The AUC was compared as a γ/(γ + β) mRNA ratio. shGADD34 and shCReP did not change the γ/(γ + β) ratio relative to the shCTRL. Error bars express ± standard error of the mean of 3 independent experiments (3 unique donors). (G) HPLC was performed on day 20 of differentiation to assess the percentage of HbF in shCTRL-, shGADD34-, and shCReP-infected cells. Due to the differences in HbF levels at baseline among donors, the percentage of HbF is shown as fold increase over the shCTRL. Both shGADD34 and shCReP significantly enhances the percentage of HbF relative to the control. Error bars express ± standard error of the mean of 3 independent experiments (3 different donors). P values were determined using 1 sample, two-tailed t test. *P < .05; **P < .01.
Complementary pharmacologic and genetic methods augment p-eIF2α and increase HbF. (A) Human primary erythroid cells were treated with 5 μM BT and 25 μM GBZ on days 15 and 18 of differentiation. The mRNA expression for γ- and β-globin was assessed from day 7 through day 20 and the AUC was calculated for each. BT and GBZ treatments did not change the γ/(γ + β) AUC mRNA ratio relative to untreated control. The error bars represent ± standard error of the mean of 3 independent experiments assessing BT treatment and 2 independent experiments analyzing the effects of GBZ. (B) HPLC was used to determine the amount of HbA and HbF present on day 20 from lysates prepared from erythroid progenitors treated with 5 μM BT and 25 μM GBZ. Both BT and GBZ significantly increase the percentage of HbF relative to the untreated control. Error bars represent ± standard error of the mean of 3 independent experiments assessing BT treatment and 2 independent experiments analyzing GBZ-treated cells. P values were determined using an unpaired two-tailed t test. *P < .05; **P < .01. (C) Differentiating human primary erythroid progenitors were infected with a control shRNA (shCTRL) and shRNAs targeting either GADD34 (shGADD34) or CReP (shCReP) on days 12 and 13. On day 16, transcript levels were evaluated for GADD34 and CReP using quantitative PCR. Both shGADD34 and shCReP resulted in approximately 50% knockdown when normalized to the shCTRL. Error bars denote ± standard error of the mean of 3 independent experiments (3 different donors). (D) Western blot analysis shows shGADD34 and shCReP result in significant knockdown of GADD34 and CReP protein, respectively, in comparison with shCTRL. Protein lysates were taken on day 16 of differentiation after 2 infections on days 12 and 13. As a loading control, β-actin was used. (E) Protein lysates were taken on days 16 and 18 of differentiation from erythroid progenitors infected with shCTRL, shGADD34, or shCReP. Western blot analysis reveals both shGADD34 and shCReP significantly increase p-eIF2α levels compared with shCTRL. Total eIF2α and β-actin were used as loading controls. (F) The mRNA expression of γ- and β-globin was assessed throughout differentiation. The AUC was compared as a γ/(γ + β) mRNA ratio. shGADD34 and shCReP did not change the γ/(γ + β) ratio relative to the shCTRL. Error bars express ± standard error of the mean of 3 independent experiments (3 unique donors). (G) HPLC was performed on day 20 of differentiation to assess the percentage of HbF in shCTRL-, shGADD34-, and shCReP-infected cells. Due to the differences in HbF levels at baseline among donors, the percentage of HbF is shown as fold increase over the shCTRL. Both shGADD34 and shCReP significantly enhances the percentage of HbF relative to the control. Error bars express ± standard error of the mean of 3 independent experiments (3 different donors). P values were determined using 1 sample, two-tailed t test. *P < .05; **P < .01.
Because pharmacologic tools can have off-target effects, especially at higher concentrations, we sought to substantiate our results with genetic modulation of the pathway. We used shRNAs targeting GADD34 and CReP to determine if increased p-eIF2α was sufficient to induce HbF without increasing the γ/(γ + β) mRNA ratio. Primary erythroid progenitors were infected with the virus containing a control or gene targeting shRNA on days 12 and 13 of culture. We achieved an average of 85% to 90% infection efficiency (supplemental Figure 3A-B), eliminating the need for further manipulation of the cells via antibiotic selection or cell sorting. We first confirmed gene knockdown by analyzing the mRNA percent remaining relative to the shRNA control on day 16 and found that both shRNAs targeting GADD34 and CReP caused approximately 50% knockdown (Figure 5C). GADD34 and CReP protein analyzed by western blot at the same time point revealed a much greater degree of knockdown (Figure 5D). This suggests that reducing GADD34 and CReP inhibited protein synthesis, which contributed to the enhanced protein knockdown observed. Importantly, then we confirmed that GADD34 and CReP knockdown significantly increased p-eIF2α relative to the shRNA control on days 16 and 18 (Figure 5E). We also found that reducing GADD34 and CReP in these cells did not increase canonical downstream ISR signaling, similar to the effects observed with Sal treatment (supplemental Figure 4A).
Next, we analyzed the effects of GADD34 and CReP knockdown on globin production. Consistent knockdown of these proteins over many days resulted in reduced mRNA expression of both γ- and β-globin (supplemental Figure 4B), which was not previously observed with Sal treatment. However, this reduction was proportional, so that neither GADD34 nor CReP knockdown changed the γ/(γ + β) mRNA ratio relative to the shRNA control during differentiation (Figure 5F). Then we evaluated HbF production using HPLC and found that both GADD34 and CReP knockdown caused a 2.5-fold increase in the percentage of HbF (Figure 5G). These results confirm that increasing p-eIF2α is sufficient to post-transcriptionally upregulate HbF in primary erythroid cells.
Combining salubrinal with activators of γ-globin transcription enhances HbF induction
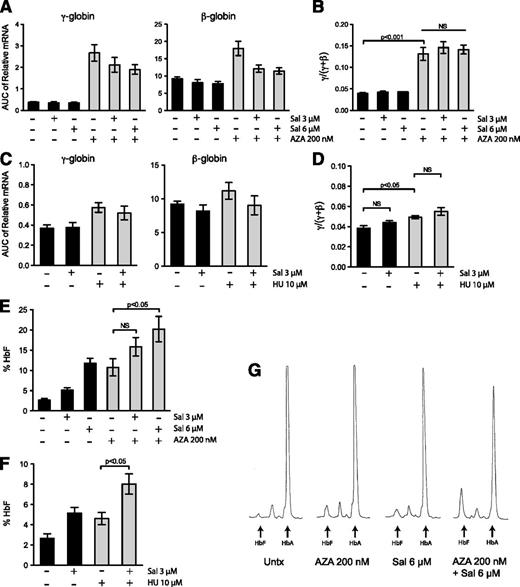
Because Sal increases HbF production post-transcriptionally, we tested whether this unique mechanism of induction could enhance the HbF induction by clinically relevant compounds that increase γ-globin mRNA levels. We chose to use AZA because it is an effective and robust inducer of γ-globin gene expression in both SCD11 and β-thal12 patients, and we chose to use HU because it is the Food and Drug Administration-approved drug for SCD, although it produces a more modest increase in HbF.13 Primary erythroid progenitors were treated every day from days 9 through 16 (with 200 nM AZA) and once on day 9 (with 10 μM HU). AZA and HU increased γ-globin mRNA expression (Figure 6A,C) to a similar extent as reported in previous in vitro differentiation studies.19,42 When Sal was combined with AZA or HU, the combination did not change cellular differentiation (supplemental Figure 5A-B) or significantly increase the γ/(γ + β) mRNA ratio relative to either single agent alone (Figure 6B,D). We then compared these results to the amount of HbF protein evaluated by HPLC on day 20. AZA alone increased the percent HbF from 2.7% to 10.8%. When 200 nM AZA was combined with 3 μM and 6 μM Sal, the percentage of HbF increased to 15.9% and 20.2%, respectively (Figure 6E and supplemental Table 1). Similarly, 10 μM HU alone increased HbF from 2.7% to 4.6%, but co-treatment with 3 μM Sal increased HbF to 8% (Figure 6F). Representative HPLC traces reveal this increased effect is due to an additive increase in HbF protein, as well as a decrease in HbA (Figure 6G). Taken together, these results indicate that the novel method of HbF induction by Sal additively enhances the effect of transcriptional activators of γ-globin gene expression.
Combination of salubrinal with AZA or HU significantly enhances HbF production post-transcriptionally in differentiating primary erythroid cells. (A) Erythroid progenitors were treated with 3 μM salubrinal (Sal), 6 μM Sal, 200 nM AZA, or in combination with Sal and AZA at these doses. The mRNA expression of γ- and β-globin was evaluated from days 7 through 20 of the culture and the AUC was calculated for each transcript. AZA treatment increases globin expression, whereas AZA in combination with Sal treatment reduces this enhancement. Globin gene expression still remains elevated in the combination-treated cells relative to the untreated control. Error bars represent ± standard error of the mean of 4 independent experiments (3 unique donors). (B) For all treatments, the AUCs were compared as a γ/(γ + β) mRNA ratio. AZA (200 nM) significantly enhances the γ/(γ + β) ratio, and the combination of Sal and AZA treatments did not change this induction. Error bars express ± standard error of the mean of 4 independent experiments (3 unique donors). P values were calculated using an unpaired two-tailed t test. (C) Erythroid progenitors were treated with 10 μM HU or 3 μM Sal alone or in combination. The mRNA expression of γ- and β-globin was evaluated from day 7 through day 20 of the culture and the AUC was calculated for each transcript. Error bars represent ± standard error of the mean of 3 independent experiments (2 different donors). (D) The AUCs were compared as a γ/(γ + β) mRNA ratio. HU treatment increases the γ/(γ + β) ratio but the combination with Sal did not significantly change this enhancement. Error bars represent ± standard error of the mean of 3 independent experiments (2 different donors). P values were calculated using an unpaired two-tailed t test. (E) The percentage of HbF was determined by HPLC on day 20 for cells treated with AZA and Sal alone or in combination. The combination of AZA and Sal dose dependently increases the percentage of HbF. Although the combination of 200 nM AZA and 3 μM Sal did not result in a significant enhancement of HbF relative to AZA alone, the combination with 6 μM Sal was significant. Error bars represent ± standard error of the mean of 4 independent experiments using 3 unique donors. P values were calculated using an unpaired two-tailed t test relative to the 200 nM AZA treatment. (F) HPLC was performed on day 20 and the percentage of HbF was calculated for cells treated with 10 μM HU and 3 μM Sal alone or in combination. The combination of HU and Sal greatly enhances the percentage of HbF relative to HU treatment alone. Error bars represent ± standard error of the mean of 3 independent experiments using 2 different donors. P values were calculated using an unpaired two-tailed t test relative to the 10 μM HU treatment. (G) HPLC was used to evaluate the proportions of HbF and HbA at the end of the differentiation process. These representative HPLC traces show the combination of 200 nM AZA and 6 μM Sal additively increases HbF while also decreasing HbA.
Combination of salubrinal with AZA or HU significantly enhances HbF production post-transcriptionally in differentiating primary erythroid cells. (A) Erythroid progenitors were treated with 3 μM salubrinal (Sal), 6 μM Sal, 200 nM AZA, or in combination with Sal and AZA at these doses. The mRNA expression of γ- and β-globin was evaluated from days 7 through 20 of the culture and the AUC was calculated for each transcript. AZA treatment increases globin expression, whereas AZA in combination with Sal treatment reduces this enhancement. Globin gene expression still remains elevated in the combination-treated cells relative to the untreated control. Error bars represent ± standard error of the mean of 4 independent experiments (3 unique donors). (B) For all treatments, the AUCs were compared as a γ/(γ + β) mRNA ratio. AZA (200 nM) significantly enhances the γ/(γ + β) ratio, and the combination of Sal and AZA treatments did not change this induction. Error bars express ± standard error of the mean of 4 independent experiments (3 unique donors). P values were calculated using an unpaired two-tailed t test. (C) Erythroid progenitors were treated with 10 μM HU or 3 μM Sal alone or in combination. The mRNA expression of γ- and β-globin was evaluated from day 7 through day 20 of the culture and the AUC was calculated for each transcript. Error bars represent ± standard error of the mean of 3 independent experiments (2 different donors). (D) The AUCs were compared as a γ/(γ + β) mRNA ratio. HU treatment increases the γ/(γ + β) ratio but the combination with Sal did not significantly change this enhancement. Error bars represent ± standard error of the mean of 3 independent experiments (2 different donors). P values were calculated using an unpaired two-tailed t test. (E) The percentage of HbF was determined by HPLC on day 20 for cells treated with AZA and Sal alone or in combination. The combination of AZA and Sal dose dependently increases the percentage of HbF. Although the combination of 200 nM AZA and 3 μM Sal did not result in a significant enhancement of HbF relative to AZA alone, the combination with 6 μM Sal was significant. Error bars represent ± standard error of the mean of 4 independent experiments using 3 unique donors. P values were calculated using an unpaired two-tailed t test relative to the 200 nM AZA treatment. (F) HPLC was performed on day 20 and the percentage of HbF was calculated for cells treated with 10 μM HU and 3 μM Sal alone or in combination. The combination of HU and Sal greatly enhances the percentage of HbF relative to HU treatment alone. Error bars represent ± standard error of the mean of 3 independent experiments using 2 different donors. P values were calculated using an unpaired two-tailed t test relative to the 10 μM HU treatment. (G) HPLC was used to evaluate the proportions of HbF and HbA at the end of the differentiation process. These representative HPLC traces show the combination of 200 nM AZA and 6 μM Sal additively increases HbF while also decreasing HbA.
Discussion
SCD and β-thal continue to cause significant morbidity and mortality on a global scale.43 Because a large proportion of hemoglobinopathy patients reside in countries that lack advanced facilities for diagnosis and management of these diseases,44 these patients require improved treatments that are safe and attainable worldwide. Considering these constraints, our goal has been to develop novel strategies to induce HbF pharmacologically, which have the greatest potential to be inexpensive and effective therapies. Although most previous studies have focused on increasing γ-globin gene expression as an approach to induce HbF, there is also evidence that HbF may be post-transcriptionally regulated.16-19 However, no molecular pathway has been previously implicated in this mode of regulation. Therefore, we chose to focus on better understanding this level of HbF production with the intention of discovering new pharmacologic targets.
In this study, we show that increasing p-eIF2α can differentially regulate HbF and HbA post-transcriptionally. We used Sal, an inhibitor of eIF2α dephosphorylation, as a tool compound to increase p-eIF2α in human primary erythroid cells. Sal did not alter cell growth or differentiation in our in vitro system when treated early or late in the differentiation process. We found that Sal did not significantly change γ- or β-globin mRNA expression but dose-dependently increased the percentage of HbF, revealing a post-transcriptional HbF induction mechanism. Furthermore, we supported these findings by testing other ISR pathway activating agents, BT and GBZ, and by genetically reducing GADD34 and CReP expression. These results combined confirm the involvement of p-eIF2α in the induction of HbF.
A surprising result was the lack of robust downstream pathway activation, despite elevated p-eIF2α produced by Sal treatment and our knockdown studies. A plausible explanation for this finding is that the ISR pathway is already activated during erythroid differentiation, as previously shown in the murine system.45 It is also known that ATF4 is a short-lived and unstable protein that is rapidly degraded by the proteasome.46 Therefore, another possible explanation is that a transient increase in ATF4 may have been present but not captured by our time course. Alternatively, previous studies have reported a similar disconnect in which specific stresses increased p-eIF2α but did not increase ATF4 protein.47 This finding, as well as our results, suggest that stress-specific or general regulation of ATF4 may be more complex in human erythroid cells. Our studies also revealed that ISR signaling trended down after many days of Sal treatment or GADD34 and CReP knockdown. Consistent with this, Zhang et al48 found downstream ISR signaling was decreased, despite increased p-eIF2α after multiple days of Sal administration in vivo. These results probably reflect reduced protein translation to an extent that eIF2 can no longer be recycled for more rounds of translation initiation. Even proteins that are selectively translated when the concentration of ternary complex is low will stop being translated eventually.
Our results also revealed differences in the degree of eIF2α phosphorylation and how this affected globin mRNA expression. Although both Sal treatment and shRNA studies did not change the γ/(γ + β) mRNA ratio, GADD34 and CReP knockdown reduced total globin mRNA expression, whereas Sal did not. This difference was probably due to the dramatic increase in p-eIF2α evident with knockdown of GADD34 and CReP relative to the more subtle changes in p-eIF2α elicited by Sal treatment. These results highlight an important feature of the p-eIF2α pathway. Slight activation is beneficial because temporarily reducing translation allows the cell to save energy, selectively translate proteins to mediate cell survival, and restore homeostasis.49 In the presence of more intense cell stress, translation is decreased to such an extent that this adaptive cell response is tipped to cell destruction and eventual apoptosis. Although we use GADD34 and CReP knockdown in this study to illustrate the involvement of p-eIF2α in the post-transcriptional induction of HbF, the chronic stress of removing these 2 important proteins is ultimately detrimental and globin expression is reduced. In contrast, pharmacologically perturbing this pathway with Sal was able to induce HbF and increase p-eIF2α to an extent that was well tolerated. This suggests that targeting this pathway therapeutically could be achievable.
Our findings and previous reports provide good rationale for increasing p-eIF2α with Sal, or a similar agent, to treat β-hemoglobinopathy patients. Recently, Suragani et al45 found that treating erythroid precursors from β-thal mice with Sal augmented p-eIF2α, reduced globin translation, and decreased the accumulation of insoluble α-globin that causes oxidative stress and apoptosis. Our current work revealed another potential therapeutic benefit of increasing p-eIF2α because it also caused induction of HbF. Elevated HbF could reduce the globin chain imbalance while simultaneously producing functional hemoglobin to alleviate the anemia in β-thal. Moreover, we found that Sal reduced HbA while increasing HbF, which could be applied to decrease the amount of mutated β-chain and reduce hemoglobin polymerization in SCD. Although pharmacokinetic studies have been performed in mouse models with Sal,48 more work will be required to find a suitable agent that is more potent and one that displays enhanced solubility to achieve clinical applicability.
Most importantly, we found that increasing p-eIF2α caused HbF to be induced through a post-transcriptional mechanism. Although the exact post-transcriptional mechanism is yet to be determined, we are actively investigating possibilities to explain these results. It is unlikely that differences in globin mRNA stability can explain the increased HbF because Sal did not change γ- or β-globin steady-state mRNA levels. It is more likely that mRNA cellular location and resulting access to the translation machinery or differences in globin mRNA translation efficiency could explain our reported findings. Additionally, enhanced p-eIF2α has been shown to increase the protein chaperone reserve in the cell to assist protein folding during stress.50 It is also possible that γ- and β-globin protein stability may be different in a dynamically changing protein environment, especially if it is assisted by different protein chaperones.
In conclusion, we found that increasing eIF2α phosphorylation, by 3 unique pharmacologic agents and shRNA targeting 2 independent proteins, caused an increase in HbF. For the first time, this revealed a connection between p-eIF2α and a post-transcriptional mechanism of HbF induction. Notably, this pathway is druggable and can be activated without altering erythroid proliferation, differentiation, or total hemoglobin content. We also demonstrated that this novel post-transcriptional mechanism of HbF induction could be combined with transcriptional activation of γ-globin gene expression to produce an additive effect. In the future, utilization of transcriptional and post-transcriptional mechanisms of HbF induction may provide an opportunity for combination therapy to achieve therapeutic HbF levels at reduced doses, thereby reducing toxicity. Our results suggest that augmenting p-eIF2α, either alone or in combination with known HbF inducers, holds promise to be an effective therapeutic strategy for β-hemoglobinopathies.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Bertal Aktas for the generous gift of BTdCPU, Dr Patrick Gallagher for kindly supplying CD34+ cells, Dr Lionel Lewis and Bernie Beaulieu for assistance with hemoglobin (Hb) HPLC, Dr Elizabeth McCoy and Dr Alexei Kisselev for guidance regarding polysome profiling, and Dr Jane-Jane Chen, Dr Rodwell Mabaera, Dr Rachel West, Dr Elizabeth Macari, and Emily Schaeffer for valuable discussions.
This work was supported by grants from the National Institutes of Health National Heart Lung and Blood Institute (HL73442) (C.H.L.) and the National Institutes of Health National Institutes of Diabetes and Digestive and Kidney Diseases (F30DK094540) (C.K.H.), and by unrestricted research funding from the Knights of the York Cross of Honour and the Royal Order of Scotland, Masonic charitable organizations.
Authorship
Contribution: C.K.H. and C.H.L. designed the research; C.K.H. performed the research; C.H.L. oversaw the research; and C.K.H. and C.H.L. analyzed and interpreted the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christopher H. Lowrey, Section of Hematology/Oncology, Dartmouth-Hitchcock Medical Center, One Medical Center Dr, Lebanon, NH 03756, e-mail: christopher.h.lowrey@dartmouth.edu.