Abstract
Despite advancements in diagnosis and treatment modalities for hematological malignancies, presently, there are no reliable blood tests for disease prognosis or prediction of patient’s response to particular treatments. Identification of such biomarkers for diagnostic or therapeutic response in clinical monitoring of hematologic diseases such as Chronic Myeloid Leukemia (CML) would be very useful.
The ability to identify biomarkers in body fluids (i.e., plasma or serum) will not only involve minimally invasive procedures, but also more importantly, the potential for monitoring disease progression and effective response to therapy in relatively early stages. In this study, global proteome analysis of serum, plasma, and bone marrow samples from 28 patients with newly diagnosed chronic–phase CML were analyzed by using expression proteomics technology (label free quantitative liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS). The goal was to evaluate for early treatment response and discovery disease–specific /disease-associated proteins for prognostic monitoring and patient’s response to therapy at protein level.
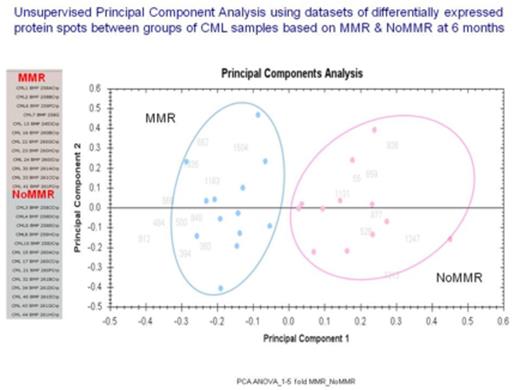
We identified a panel of 15 differentially expressed proteins (> 2- °- fold change, p< 0.001) to accurately discriminate between CML patients that achieved Major Molecular Response (MMR) vs. No-Major Molecular Response (NoMMR) at 6 months (Figure 1 & Table 1). Six (6) of the 15 identified proteins were filtered and mapped as potential biomarkers using Ingenuity Pathway Analysis. Among the identified proteins implicated in hematological diseases include Group-specific component (vitamin D binding protein), haptoglobin and vitronectin. Others were inter-alpha-trypsin inhibitor heavy chain 1, leucine-rich alpha-2-glycoprotein 1 and metallophosphoesterase 1. We also observed most of these proteins to be located in extracellular space, acts as transporters and have been implicated as markers of hematological disease and inflammatory response. Our data indicates that multivariate analysis of quantitative proteome data can potentially be useful as a means of unsupervised artificial intelligence algorithm for classification and stratification of clinical CML patients.
List of differentially expressed proteins between CML samples that achieved major Molecular response versus No major Molecular response (Proteins were identified using SynaptG2 coupled with Nano LC (Waters)
 |
 |
Our results highlight the power of proteomics in the discovery of biomarkers for more accurate prediction of prognosis and monitoring treatment response in CML patients. These proteins might be valuable, once validated, to complement the currently existing parameters for reliable and objective prediction of disease progression, monitoring treatment response and clinical outcome of CML patients.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal