Abstract
Imatinib resistance in CML is commonly associated with point mutations within the BCR-ABL1 kinase domain (KD). Mutation analysis can therefore guide selection of the most appropriate alternative therapy after imatinib failure, as several tyrosine kinase inhibitors (TKIs) with different resistance profiles are available as next-line therapy. Sanger sequencing is the most common mutation analysis technique used, but has limited sensitivity and cannot distinguish between mutations that are on the same (compound) or different molecules (polyclonal). This is becoming increasingly important clinically because BCR-ABL1 compound mutants were predicted from preclinical studies to cause resistance to ponatinib.
Using PCR amplicons, several groups have recently reported that 65-85% of multiple mutations in CML patients with TKI resistance are compound, indicating the potential for significant clinical resistance to ponatinib (Khorashad Blood 2013; Soverini Blood 2013). Perhaps surprisingly, in the majority of cases, the mutations were found to be both components of compound mutations and present as individual mutations in the same patient. This suggests that the same nucleotide substitution occurred independently multiple times within individual patients. This has been reported for JAK2 in a study of myeloproliferative disorders. However, the complexity observed in CML is difficult to explain phylogenetically, and the dynamics of these compound mutant populations in relation to TKI treatment in vivo appear unpredictable.
It is widely reported that PCR-mediated recombination between highly similar sequences frequently causes formation of chimeras which can greatly increase estimates of clonal diversity. Using mixtures of plasmids with known BCR-ABL1 KD mutations, we aimed to determine the frequency of PCR-mediated recombination which may mimic BCR-ABL1 compound mutations.
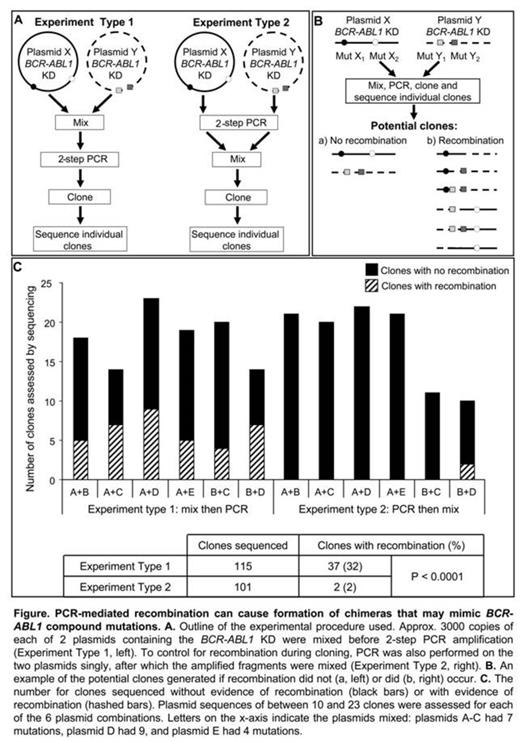
PCR and cloning procedures previously used to detect compound mutations in CML patients were replicated. To examine the potential for PCR-mediated recombination, 2 different types of cloning experiments were performed using 5 plasmids known to harbor 4 to 9 BCR-ABL1 KD mutations each: 1) two plasmids were mixed and then 2-step nested PCR amplification was performed prior to cloning, and 2) 2-step nested PCR was performed on these plasmids singly and the amplified fragments were then mixed before cloning, Figure A. PCR was performed using Expand Long Template PCR System (Roche) and amplified fragments were cloned into E. Coli strain JM109 to minimize E. Coli-mediated recombination and repair of heterologous DNA. Clones were Sanger sequenced to reveal the sequence of individual amplified fragments. This process was replicated with 6 different combinations of plasmids.
If PCR recombination had not occurred, sequences of individual clones would show specific variations that originated from one or the other original plasmid. Conversely, if PCR recombination had occurred, individual clones would have specific variations originating from both of the mixed plasmids, Figure B.
When plasmids were mixed before PCR (Experiment Type 1), clones of all 6 plasmid mixtures showed evidence of recombination, some had even undergone 2 or 3 recombination events. 26-50% of the clones had patterns of recombination similar to that outlined in Figure Bb. New configurations of compound mutations in individual amplicons were observed. The new configurations of mutations resulted in greater genetic diversity than in the original mixture, and were similar to those reported in studies of CML patients. In comparison, when the singly amplified plasmids were mixed and cloned (Experiment Type 2), recombination was observed in clones of 1 of the 6 plasmid mixtures. In total, 32% (37/115) of clones of Experiment Type 1 were chimeras, compared to 2% (2/101; 0-20%) of clones of Experiment Type 2, p<.0001, Figure C.
Our study provides evidence that PCR can mediate the formation of chimeras that may mimic BCR-ABL1 compound mutations. This could lead to imprecision in estimation of clonal diversity and compound mutation frequency. It highlights the need for caution when interpreting results using current procedures, and the need for new techniques to enable assessment of the clinical impact of BCR-ABL1 compound mutations on patient response to third-generation TKIs.
Yeung:Novartis: Honoraria, Research Funding; BMS: Honoraria. Hughes:Novartis: Consultancy, Honoraria, Research Funding; Bristol Myers Squibb: Consultancy, Honoraria, Research Funding; Ariad: Consultancy, Honoraria; CSL: Research Funding. Branford:Novartis: Honoraria, Research Funding; BMS: Honoraria, Research Funding; Ariad: Honoraria, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal