Abstract
Myc-interacting zinc finger protein 1 (Miz-1; Zbtb17) is a BTB/POZ (POZ) domain transcription factor expressed in all mammalian tissues. The POZ domain is required for Miz-1 function and favors a stable association with chromatin. As a binding partner of the oncoprotein c-Myc, Miz-1 can modulate the expression of c-Myc target genes (e.g. p21Waf1, p15ink4b). c-Myc is frequently associated with a variety of leukemia and lymphoma. In particular, c-Myc takes part in the t(8;14) chromosomal translocation where it is placed under the control of transcriptional regulatory elements of the immunoglobulin heavy chain (IgH) locus. This chromosomal translocation is a hallmark of patients with Burkitt-type B-cell lymphoma (BL). Since epigenetic deregulation is often associated with tumor development, we wished to determine whether (i) Miz-1 is involved in the development of c-Myc-dependent B-cell lymphoma and whether (ii) Miz-1 influences c-Myc dependent chromatin and gene regulation.
We are using the Eµ-Myc mouse model to generate Burkitt type c-Myc dependent lymphomas to study the implication of Miz-1 in a c-Myc driven process of malignant transformation. Mice carrying the Eµ-Myc transgene (c-Myc gene placed under the control of Eµ enhancer of the IgH locus) overexpress c-Myc in lymphoid cells and develop a disease similar to Burkitt's lymphoma. Mice that express a conditional non-functional Miz-1 allele lacking the part coding for the POZ domain in B cells (Mb1-Cre-Miz-1DPOZ/flox mice, hereafter called DPOZ mice) were crossed with Eµ-Myc transgenic mice. Incidence and latency periods of the development of tumors in these animals were compared to Eµ-Myc animals that express a functional Miz-1 protein. Pre-cancerous mice of all genotypes were also investigated to evaluate the influence of Miz-1 during the development of the c-Myc dependant lymphomas. To address the role of Miz-1 in the deregulation of chromatin associated with c-Myc overexpression, chromatin immunoprecipitation coupled to high throughput sequencing (ChIP-seq) was performed on B-lymphoma and pre-tumoral B-cells and compared to normal B-cells. Binding to chromatin of Miz-1, c-Myc as well as histone marks will be assessed. RNA from each cell type will also be submitted to high throughput sequencing (RNA-seq) to correlate the binding of transcription factors with gene expression.
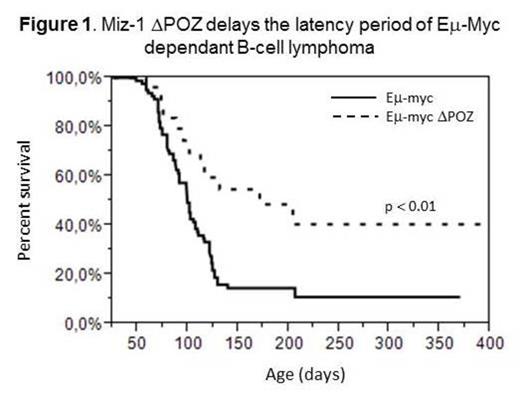
Eµ-Myc mice that express the non-functional Miz-1 protein (DPOZ) in B-cells develop lymphoma later than Eµ-Myc mice that express a normal, functional Miz-1 (Figure 1). Tumors that grow in Eµ-Myc/DPOZ animals are phenotypically similar, but are smaller compared to those developing in Eµ-Myc mice. Additionally, blood analysis of sick animals reveal a lower amount of Large Unstained Cells (LUC) when Miz-1 is mutated. This suggests that Myc driven lymphoma and leukemia is impaired in the absence of a functional Miz-1 protein. Accordingly, in 40 days-old pre-tumoral Eµ-Myc/DPOZ animals compared to Eµ-Myc mice with wild type Miz-1, less pre-B and/or pro-B cells were observed in lymphoid organs and little or no LUC were present in the blood, suggesting that a c-Myc driven B-cell lymphoma develops with a longer latency period and progresses at a slower rate in the absence of a non-functional Miz-1 protein.
Our preliminary data indicate that Miz-1 is involved in the development of aggressive c-Myc driven B-cell lymphoma. To test the potency of Eµ-Myc/DPOZ tumor cells to initiate B-cell lymphoma, we will transplant these tumor cells into immune-deficient mice and follow the development of the transplanted cells. Also, to test whether Miz-1 would be a suitable therapeutic target for future leukemia and lymphoma treatment, we will use a conditional model (ROSA-CreER-Miz-1DPOZ/flox) where the POZ domain coding sequences of Miz-1 can be deleted after tamoxifen treatment after tumor initiation in Eµ-Myc animals. To better understand the mechanisms by which Miz-1 influences c-Myc in the process of lymphomagenesis, we are performing high throughput genomic analyses to identify genes that are differentially regulated in Eµ-Myc lymphomas when Miz-1 is functional or deficient. These analyzes should demonstrate whether a c-Myc/Miz-1 complex is required to malignantly transform B-cells or whether c-Myc and Miz-1 act independently in the development of B-cell lymphoma.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal