Abstract
Cell fate and lineage specification have been thought to be largely regulated at the level of transcription. However, regulation of differentiation at the transcriptional level alone does not appear to explain all hematopoietic cell fate decisions, suggesting the presence of other as-yet-unknown mechanisms. EBF1 is critical transcription factors in B-lymphopoiesis. Ectopic EBF1 expression promotes B cell development in multipotent progenitor cell (MPP). EBF1is deleted and downregulated in different B cell malignancy like some ALLs and Hodgkin lymphoma.
MicroRNAs are noncoding RNAs that are vital to many cell functions and that post-transcriptionally repress target mRNAs. Each miRNA is thought to have several target mRNAs, and computational predictions suggest that more than a third of all human genes are targets of miRNAs. However, in cell fate determination, it has been thought to act as only a fine tuner.
Recently we reported that miR-126 drives MLL-AF4 ALL cells, exhibiting both myloid and B cell surface markers, towards B cell without upregulating of EBF1, E2A, and PAX5, which are critical transcriptional factors in B cell lineage commitment. Moreover, miR-126 could induce partial B lymphopoiesis in EBF1 deficient hematopoietic cells (HPCs) (Okuyama.K et al. PNAS).
In the present study, in order to challenge the canonical notion that cell fate is governed solely by transcriptional factors, we investigated whether miRNA have full potentiality to induce B cell development in EBF1 deficiency.
We introduce several different miRNA in EBF1-/-hematopoietic progenitor (Lin-) cells and cultured them on TSt-4 stromal cells in IMDM medium containing stem cell factor, IL-7, and Flt3 ligand (10 ng/mL each). Then comprehensive Gene-Expression analysis, genomic PCR for VDJ recombination analysis and flowcytometric analysis for B-cell lineage markers were performed.
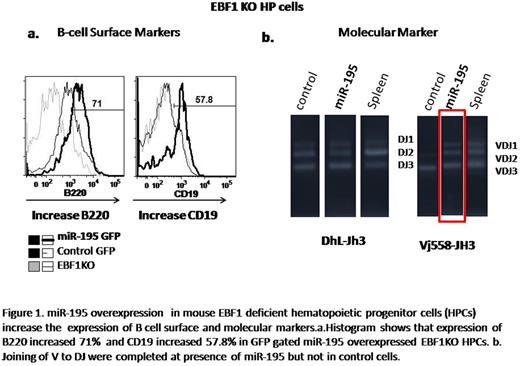
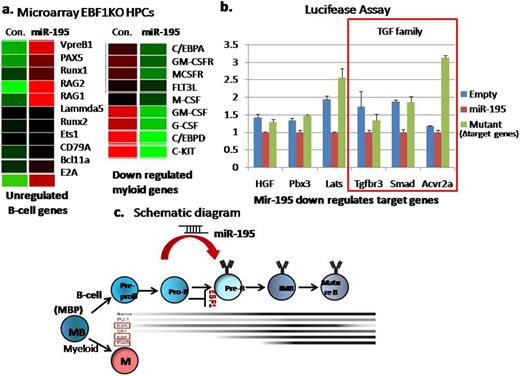
Among the miRNA we analyzed, miR-195 induced expression of CD19, an established B lineage marker, and upregulated of B220 , B cell associated marker, comparing with control vector expressed EBF1 KO HPCs (Figure 1a). Then we investigated the molecular marker of B committed cells,“ VDJ recombination”. In miR-195 transduced EBF1KO cells, V to DJ recombination were completed, while the control was not (Figure 1b). Based on the results obtained from surface and molecular markers, miR-195 transduced EBF1KO cells were identified as B committed cells suggesting that miR-195 can rescue B lymphopoiesis in EBF1 deficiency. Finally our microarray data shows that overexpression of miR-195 in EBF1 KO cells upregulates B cell lineage commitment genes such as Pax5, Bcl11a, VpreB1, Cd79a, lamda5 and E2A which are downsteam targets of EBF1 and downregulates the myeloid lineage specific genes which are also antagonized by EBF1 in B cell lineage specification (Figure 2a).
Next in order to clarify its mechanism, we studied the target genes of miR-195. Tgfβr3, Smad7, Acvr2a, Lats, HGF, and Pbx3 were identified as the targets of miR-195 by luciferase assay in 293T cells (Figure 2b). TGF beta family and the pathway, has been reported to suppress B lymphopoiesis. We hypothesized that TGF beta family genes such as Tgfβr3, Smad7, and Acvr2a, are responsible for transcription factor independent B cell differentiation by miR-195. Further analysis is now under investigation.
MiR-195 induces CD19 expression, completion of VDJ recombination and upregulation of B cell related genes in EBF1-/-HPCs while neither PAX5 nor E2A are able to upregualte B cell related genes or induce CD19 expression in the absence of EBF1. It suggests that miR-195 is more potent than E2A and Pax5 in this system and acts more than a fine tuner in B cell lineage specification. In the canonical notion, cell fate is determined solely by transcriptional factors. Our study challenges this notion, proposing that miRNA might have potential to be an alternative of transcriptional factors in some condition (Figure 2c). As we all know, impaired differentiation by deregulation of transcriptional factors is one of “hit” to leukemia development. Accordingly miRNA can potentially rescue this deregulation to become promising therapeutic target.
Ando: Alexion: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal