Abstract
Many reports state that hematopoietic malignancies mostly result from somatic mutations in HSCs in the bone marrow. Somatic mutations of spliceosomal gene such as SF3B1, U2AF1 and SRSF2 have been widely described in myelodysplastic syndrome (MDS). Some studies presented that MDS patient with splicing factor mutations influence the clinical outcomes. However, the clinical significances for the treatment of hypomethylating agents (HMA) in splicing factor mutation were not reported. Therefore, this study investigated the influences of the SF3B1, U2AF1 and SRSF2 splice gene mutation in MDS patients who received the HMAs.
MDS harboring ring sideroblast and association with somatic spliceosomal gene mutation was well demonstrated but, comparatively rare and showed good prognosis. So, we excluded MDS harboring ring sideroblast in this study. The study cohort of 133 MDS patients without harboring ring sideroblast was examined for somatic mutations in SF3B1, U2AF1 and SRSF2 splicing gene using direct sequencing method and 59 out of 133 patients received the treatment of HMAs (43 of Azacitidine and 16 of decitabine) for the treatment of MDS. Using the international prognostic scoring system(IPSS), the treatment indications for the HMA were as follows, 1) inermediate-1 with anemia and no response for the treatment of erythropoietin, 2) intermediate-1 with anemia accompanying other cytopenia ( neutrophil <1,000/uL or PLT <100,000/uL), 3) intermediate-2 or high risk. The response analysis was followed the modified IWG MDS response criteria.
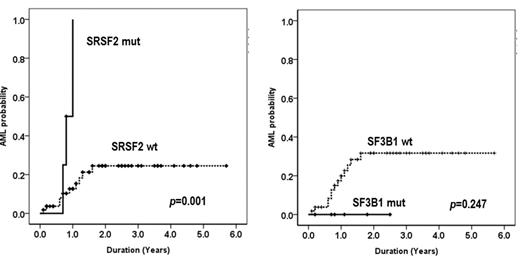
In 59 patients, mutations in K700E of SF3B1; S34T, S34P or Q157P of U2AF1; P95H or P95R of SRSF2 were found in 6 (10.2%), 7 (11.8%), and 4 (6.8%) patients, respectively. The 17 patients were observed any mutation (SF3B1, U2AF1 or SRSF2) in 59 patients. We compared the clinical features, treatment responses and survivals according to the somatic mutations of spliceosomal gene vs wild type (WT) in each mutation. The disease composition of 59 patients was like as follows; 1 of MDS with del(5q), 6 of RCUD, 24 of RCMD, 9 of RAEB-1, 19 of RAEB-2. In the clinical features, lower risk (according to IPSS, WPSS and revised-IPSS) patients was included in the group with SF3B1 mutation (P<0.05). The hematologic improvement or more response for the HMA was observed in 33% vs 47% in SF3B1 mutation vs WT, 29% vs 48% in U2AF1 and 75% vs 44% in SRSF2, respectively. There was no difference in the response rates for the HMA therapy according to the mutation or wild type (P>0.05). Overall survival did not show the statistical differences in each mutation (P>0.05). The leukemia free survival in patients with SRSF2 mutation was inferior to the WT (p=0.001). However, anyone showed the leukemic transformation in the patients with SF3B1 mutation without statistical significance (p=0.247) (Fig. 1).
AML probability of SRSF2 and SF3B1 splicing genes in MDS patients without harboring ring sideroblast. The leukemia free survival in patients with SRSF2 mutation was inferior to the WT (p=0.001). However, no statistical significance of the leukemic transformation was observed in the patients with SF3B1 mutation (p=0.247).
AML probability of SRSF2 and SF3B1 splicing genes in MDS patients without harboring ring sideroblast. The leukemia free survival in patients with SRSF2 mutation was inferior to the WT (p=0.001). However, no statistical significance of the leukemic transformation was observed in the patients with SF3B1 mutation (p=0.247).
Our results show that mutation of SF3B1, U2AF1 and SRSF2 spliceosomal gene in MDS patients without harboring ring sideroblast did not influence the treatment response and overall survival for the HMAs. However, alteration of SRSF2 splice gene may be regarded as a risk factor of leukemic transformation. So, the patients with SRSF2 mutation treated with HMA have to consider the aggressive therapy such as allogeneic stem cell transplantation before leukemic transformation. To confirm this result, it will be needed more study for large number of patients.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal