Abstract
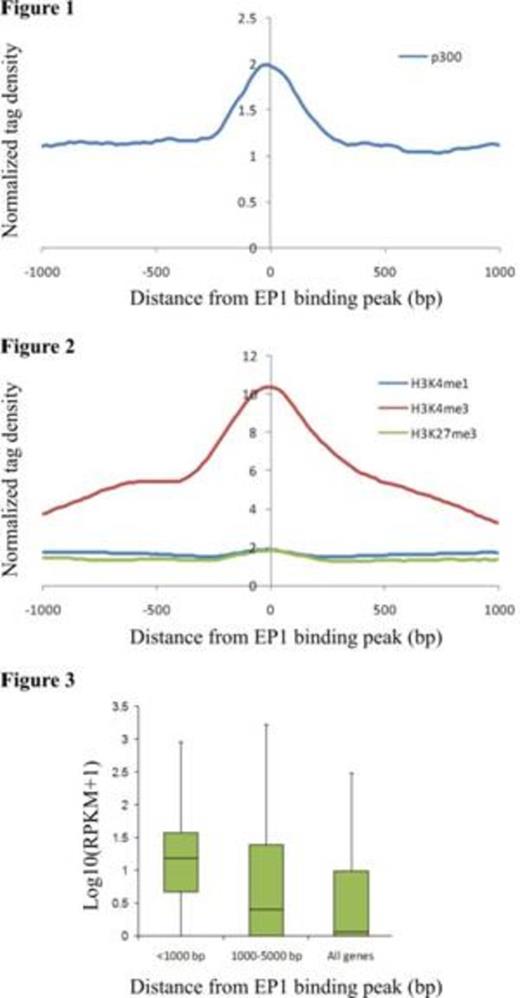
E2A-PBX1 (EP1) is a chimeric oncogenic transcription factor expressed consequent to the 1;19 chromosomal translocation in cases of acute lymphoblastic leukemia (ALL). EP1 can induce transcription of reporter genes and EP1-driven oncogenesis requires direct binding of EP1 with the transcriptional co-activator and histone acetyltransferase p300. Therefore, we hypothesized that EP1 recruits p300 and other co-activators to cis-acting regulatory elements throughout the genome thereby inducing or maintaining transcription of target genes some of which contribute to the neoplastic phenotype. Here we have used chromatin immunoprecipitation followed by next generation DNA sequencing (ChIP-seq) to identify and characterize EP1-bound sites across the genome of the t(1;19)-associated, ALL-derived cell line RCH-ACV.
ChIP was performed with an anti-FLAG antibody using sheared chromatin prepared from RCH-ACV cells that stably expressed FLAG-tagged EP1; ChIP from parent RCH-ACV cells not expressing FLAG-EP1 served as a negative control for peak calling. Parallel immunoprecipitations were performed with antibodies for p300 and the chromatin marks H3K4me3, H3K4me1 and H3K27me3. Sequencing of DNA purified from the immunoprecipitated material and of total RNA (RNA-seq) was carried out commercially by BGI whereas bioinformatic analyses were performed in-house.
In summary, our results suggest that EP1 recruits p300 and other co-activators to transcriptionally active chromatin in ALL cells. Results from studies currently underway to confirm the dependency of target gene expression and p300 recruitment upon binding of EP1 at specific binding sites will be presented.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal