Abstract
In multiple myeloma (MM), unlike normal plasma cells (PC), myeloma cells retain the self-renewing potential. Majority of medullary myeloma cells regardless over-expression of cyclins D stay in the G1 phase due to pro-apoptotic and cell cycle regulatory capacity of p53 depended axis. Nevertheless, after leukemic transformation in secondary plasma cell leukemia (PCL) or de novo in case of primary PCL, bone marrow myeloma cells become highly proliferative and even presenting as circulating plasma cells in the peripheral blood. We anticipate that complex “re-setting” of cell cycle gene coordination during leukemic transformation creates required background to restore proliferation activity and breakthrough mitotic restriction points.
The objective of our study was to define and describe complex “re-setting” of cell cycle gene coordination in MM and PCL.
In total, 7 healthy donors, 6 multiple myeloma and 7 plasma cell leukemia samples enrolled in this study. The mRNA from CD138+ cells was isolated using uMACS mRNA Isolation Kit Small scale (Miltenyi Biotech) and directly amplified and labeled using WT-Ovation™ Pico RNA Amplification System Version 1.0 plus WT-Ovation™ Exon Module Version 1.0 (NuGEN). Generated SPIA-cDNA was fragmented & labeled using Encore™ Biotin Module (NuGEN). cDNA was hybridized to Affymetrix GeneChip Human Exon 1.0 ST Arrays (Affymetrix, Santa Clara, USA). All samples were labeled and scanned in a randomized order to avoid batch effects. Gen sets, connected with cell cycle regulation (GO:0045786; GO:0045787) with all direct descendants (child terms) and regulation of apoptotic process (GO:0043065; GO:0043066), were taken for the Gene Set Enrichment Analysis (GSEA) and Gene Set Differential Coordination Analysis (GSDCA).
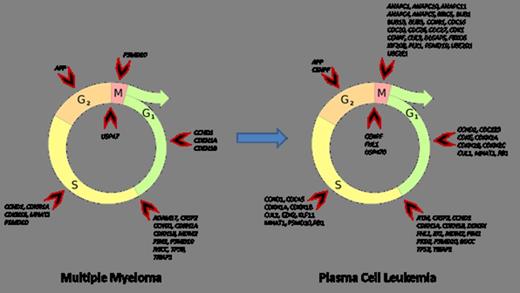
Comparing of PCL, MM and healthy donors revealed coordinating changes between regulation of mitosis (GO:0045839; GO:0045840), apoptosis (GO:0043065; GO:0043066) and cell cycle arrest (GO:0007050). These changes were relevant for both positive and negative regulation sets. Gene expression profiling of MM samples revealed affected early phases of cell cycle (G1 phase and G1/S transition). In PCL samples co-expression changes was associated with late phases of cell cycle (G2/M transition, S and M phase) together with severe alteration in early phases. The mechanisms controlling differential cell cycle coordination were based on bioinformatic analysis suggested to include alternative transcription start sites, exon skipping and shortening of 3'UTR. The probe sets covering the 3'UTR of CCND2 were for example significantly down regulated in plasma cells of MM and PCL as compared to healthy donors supporting the existence of a phenomenon observed in breast cancer where shortening of 3'UTR mRNA CCND2 confers higher mRNA stability leading to higher protein expression and more cells to enter the S phase.
Considering revealed coordination changes allow us to offer following statements. Expression of cell cycle positive regulators is in dynamic equilibrium with cell cycle negative regulators. We suppose that this equilibrium serves as a compensatory mechanism to oncogenic events. Despite compensation mechanisms activation, whole regulatory complex seems to be imbalanced by growing “oncogenic stress” during MM to PCL progression.
This study was supported by grants NT11154, NT12130, NT13190 and the EU 6th FP to MSCNET (LSHC-CT-2006-037602), the Danish Cancer Society, the Danish Research Agency (#2101-07-0007) and the KE Jensen Foundation (2006-2008).
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal