Abstract
The molecular mechanisms responsible from evolution to malignant monoclonal plasmacytosis is still under investigation. The aim of this prospective study was to analyze the proteomics profile of plasma cells obtained from MGUS, SMM and symptomatic myeloma patients to be able to investigate the differences at protein level between patients with low vs high plasma cell content (PCC).
Marrow samples were collected from 30 patients newly diagnosed with Multiple Myeloma (n: 28 symptomatic and n: 1 smoldering (SMM)) and n:1 MGUS) at Ankara University Department of Hematology. Plasma cells were isolated by CD138+ selection before protein extraction. The patients were classified mainly in to three groups according to marrow PCC by flow cytometry: group1: 0-9, group 2: 10-20 and group 3: >20 %. The protein profiles of these three groups were constructed and compared via 2D gel electrophoresis by using PDQuest 8.01 analysis. Up/down regulated protein spots were identified with Matrix-assisted laser desorption/ionization mass spectroscopy(MALDI) by Peptide Mass Fingerprinting (PMF) analysis
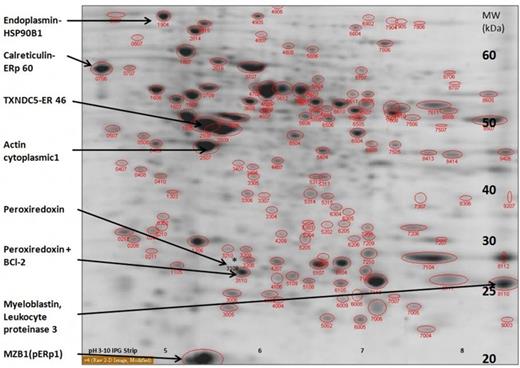
All protein spots that were detected according to PCC with PDQuest analysis are as follows: 135 spots in group 1 142 spots in group 2 and 145 spots in group 3 Among these spots, 27 spots with significant expression density difference (at least 2-fold) were detected between group 1 and 2, 36 spots between group 1 and group 3 and 28 spots between group 2 and group 3. 36 of these protein spots were were used in peptide isolation by using trypsin. PMF analyses were carried out in MALDI-TOF mass spectrometer. From these spots, eight proteins were identified by using Mascot: Endoplasmin (ERp99), Calreticulin(ERp60), MZB1(Marginal zone B and B1 cell specific protein/pERp1), Actin cytoplasmic1(ACTB), Myeloblastin (Leukocyte proteinase 3), Thioredoxin domain-containing protein 5 (TXNDC5/ERp46), Apoptosis regulator B-cell lymphoma 2 (Bcl-2) and Peroxiredoxin-4 ( Table 1, Figure 1). The remaining 28 spots are under investigation. In group 3 the density of protein spots which contain Calreticulin, MZB1, Myeloblastin and TXCDN5 increased, and which contain Actin and Endoplasmin decreased significantly (Table 1). There was a negative corelation between the Peroxiredoxin expression level and the PCC which resulted in disappearance as the percentage of PCC increased. Moreover, not only Peroxiredoxin, but also BCL-2 protein was detected only in the group with PCC >20 %. To avoid the changes that can be attributable to PC counts equal amounts of protein were analyzed from each group.
Until now there is only one published study utilizing proteomics in MM and reports 268 proteins (Chun Hua Lu et al, J Proteomics Informatics 2010). Ours is the first proteomics study comparing plasma cell proteins with PCC. The functional properties of these proteins are summarized (Table 2). High protein production and folding plus Ca changes in MM support our findings on chaperons and Ca binding protein changes. Out of these eight proteins only bcl-2, MZB1 and calreticulin were previously reported to be involved in the biology of MM.
Information on the proteins identified using Mascot and the densities of the protein spots
| MASCOT RESULT | PDQUEST RESULT | ||||||
| SSP (Sample Spot Protein) number | Protein | Score | Protein sequence coverage (%) | Mass values search /Mass values matched | Density of protein spots | ||
| 0-9 % | 10-20 % | +20% | |||||
| 0706 | Calreticulin | 102 | 32 | 10/8 | 11938 | 8046 | 17929* |
| 1904 | Endoplasmin | 100 | 14 | 10/10 | 10214* | 3034 | 5277 |
| 2011 | MZB1 | 106 | 52 | 10/7 | 16090 | 18768 | 30697* |
| 2507 | Actin cytoplasmic1 (ACTB) | 153 | 34 | 10/8 | 26183 | 28289 | 15581* |
| 2508 | TXNDC5 | 142 | 24 | 10/8 | 14905 | 14263 | 21094* |
| 3108 | Peroxiredoxin -4 | 105 | 38 | 10/7 | 6289 | 5061 | 35* |
| 3110-1 | Peroxiredoxin -4 | 123 | 58 | 25/10 | 37* | 6757*a | 9671*a |
| 3110-2 | Bcl-2 | 65 | 48 | 25/6 | |||
| 8110 | Myeloblastin | 96 | 33 | 10/5 | 10133 | 8478 | 15659* |
| MASCOT RESULT | PDQUEST RESULT | ||||||
| SSP (Sample Spot Protein) number | Protein | Score | Protein sequence coverage (%) | Mass values search /Mass values matched | Density of protein spots | ||
| 0-9 % | 10-20 % | +20% | |||||
| 0706 | Calreticulin | 102 | 32 | 10/8 | 11938 | 8046 | 17929* |
| 1904 | Endoplasmin | 100 | 14 | 10/10 | 10214* | 3034 | 5277 |
| 2011 | MZB1 | 106 | 52 | 10/7 | 16090 | 18768 | 30697* |
| 2507 | Actin cytoplasmic1 (ACTB) | 153 | 34 | 10/8 | 26183 | 28289 | 15581* |
| 2508 | TXNDC5 | 142 | 24 | 10/8 | 14905 | 14263 | 21094* |
| 3108 | Peroxiredoxin -4 | 105 | 38 | 10/7 | 6289 | 5061 | 35* |
| 3110-1 | Peroxiredoxin -4 | 123 | 58 | 25/10 | 37* | 6757*a | 9671*a |
| 3110-2 | Bcl-2 | 65 | 48 | 25/6 | |||
| 8110 | Myeloblastin | 96 | 33 | 10/5 | 10133 | 8478 | 15659* |
Comparison between 0-9 % vs 10-20 % and + 20% *P< 0.05, Comparison between 10-20 % vs +20 % aP<0.05
Chromosomal location and functional properties of the eight proteins detected ( ref: www.nextprot.org)
| Protein . | Chromosomal Location . | Chaperone . | Apoptotic process . | Oxidoreductase . | Regulation of Ca(2+) . | Cell proliferation . |
|---|---|---|---|---|---|---|
| Calreticulin | 19p13.2 | + | + | + | ||
| Endoplasmin | 12q23.3 | + | + | + | ||
| MZB1 | 5q31.2 | + | + | + | + | + |
| Actin cytoplasmic1 | 7p22.1 | + | ||||
| TXNDC5 | 6p24.3 | + | + | |||
| Peroxiredoxin -4 | Xp22.11 | + | ||||
| Bcl-2 | 18q21.33 | + | + | + | ||
| Myeloblastin | 19p13.3 | + |
| Protein . | Chromosomal Location . | Chaperone . | Apoptotic process . | Oxidoreductase . | Regulation of Ca(2+) . | Cell proliferation . |
|---|---|---|---|---|---|---|
| Calreticulin | 19p13.2 | + | + | + | ||
| Endoplasmin | 12q23.3 | + | + | + | ||
| MZB1 | 5q31.2 | + | + | + | + | + |
| Actin cytoplasmic1 | 7p22.1 | + | ||||
| TXNDC5 | 6p24.3 | + | + | |||
| Peroxiredoxin -4 | Xp22.11 | + | ||||
| Bcl-2 | 18q21.33 | + | + | + | ||
| Myeloblastin | 19p13.3 | + |
Beksac:Janssen: Honoraria, Membership on an entity’s Board of Directors or advisory committees; Celgene: Honoraria, Membership on an entity’s Board of Directors or advisory committees; Novartis: Honoraria, Membership on an entity’s Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal