Abstract
Gene expression profiling (GEP) via microarray analysis enables the measurement of expression levels for tens of thousands of genes in a single experiment, and it has been widely used in clinical practice for cancer classification, risk stratification, and treatment selection. However, results of GEP-based clinical diagnostic/prognostic tests can be highly affected by batch effects when clinical samples are processed differently (e.g. on different days, by different technicians, or using different sample protocols). Yet, this problem is rarely discussed in the literature. Understanding the role of batch effects on GEP-based conclusions is vital because GEP-based-risk treatment assignment has been used for personalized treatment to improve patients' survival: patients with low risk and favorable clinical and biological features can be treated with a less intensive, lower toxicity treatment, while patients with high risk and unfavorable features can be treated with a more aggressive, potentially more toxic therapeutic approach. Here, we investigate how sample processing discrepancies influence various GEP-based prognostic models in multiple myeloma (MM) and how to adjust for such effects during data analysis.
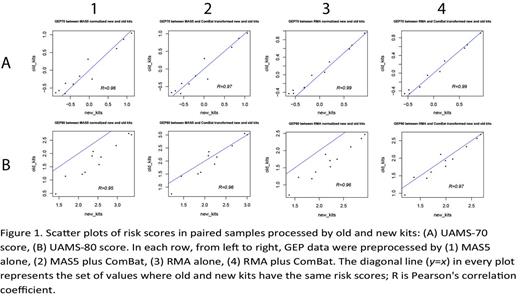
In 2009, Affymetrix discontinued their One-Cycle and Two-Cycle Target Labeling and Control Reagents (hereon referred to as the 'old' kit) and replaced it with a 3' IVT Express Kit (hereon referred to as the 'new' kit). To examine the impact of the replacement kit on GEP results, we set out to process eleven CD138-enriched patient plasma cell samples using both the new and old kits side-by-side before hybridizing them separately to the Affymetrix HG-U133 Plus 2.0 arrays. Various GEP-based MM prognostic scores, including UAMS-70, UAMS-80, UAMS-17, EMC-92, IFM-15, MRC-IX-6, and MILLENNIUM-100, were calculated and compared between the matched GEP pairs with either MAS5 or RMA normalization, with and without batch effect adjustment by ComBat (Combating Batch Effects When Combining Batches of Gene Expression Microarray Data). Both the UAMS-70 and UAMS-80 scores are based on log2 ratios between unfavorable and favorable genes regarding survival, which are self-normalized. However, with MAS5 alone, the UAMS-70 score was similar between the two kits (p-value=0.37 from paired t-test) but not for UAMS-80 score, which was significantly higher under the new kit (p-value<0.001 from paired t-test, Figure 1). Furthermore, besides UAMS-17, the score values from the EMC-92, IFM-15, MRC-IX-6, and MILLENNIUM-100 models without batch effect adjustment were all affected by the kit issue. After ComBat adjustment, variation caused by batch effects markedly reduced, and as a result, correlation increased between the prognostic scores of the two different kits. For most GEP-based MM prognostic scores, kit effect was minimized by RMA plus ComBat correction, which resulted in similar risk scores between the two kits.
For GEP-based prognostic models, it is important to check for possible batch effects which may be the end result of various causes, such as differences in sample preparation and processing protocols, like the new kit/old kit issue discussed here. We found that even for self-normalized prognostic signatures, risk scores can still sometimes be significantly different because of batch effects. So, it is essential to preprocess GEP raw data carefully, minimizing variance caused by batch effects, and adjusting any GEP-based diagnostic result accordingly, e.g. cutoff values in risk-assessment models.
Usmani:Celgene: Consultancy, Research Funding, Speakers Bureau; Onyx: Research Funding, Speakers Bureau. Barlogie:Celgene: Consultancy, Honoraria, Research Funding; Myeloma Health, LLC: Patents & Royalties.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal