Abstract
Most multiple myeloma (MM) patients will experience relapse due to the persistence of residual tumor cells, or MRD. We compared the prognostic value of traditional response criteria with MRD measured by three different methods: a sequencing-based method, termed the LymphoSIGHT™ platform, multiparameter flow cytometry (MFC) and ASO-PCR of Ig genes in a cohort of 133 uniformly-treated MM patients from the Spanish Myeloma Group trials.
Bone marrow samples were obtained from 133 patients at diagnostic and post-treatment time points on GEM clinical trials (GEM00 and GEM05). All 133 patients were either in CR or VGPR at the post-treatment time point. Using the sequencing assay, we identified clonal rearrangements of immunoglobulin (IGH-VDJ, IGH-DJ, and IGK) genes in diagnostic samples. We then assessed MRD in follow-up samples, analyzing concordance between: sequencing, MFC and ASO-PCR of Ig genes MRD results, and comparing the prognostic value of each method with traditional response criteria.
The sequencing assay detected the presence of a myeloma-specific gene rearrangement in diagnostic samples from 121 of 133 (91%) patients. We tested MRD in follow-up time points in 109 of the 121 patients.
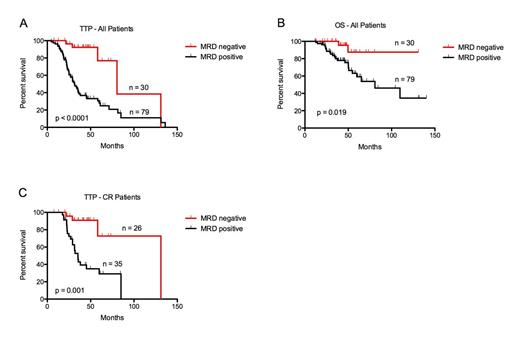
Of the 109 patients, 79 were positive by sequencing at MRD levels of 10-5 or higher and 30 were MRD negative. The Time to Tumor Progression (TTP) and Overall Survival (OS) were significantly longer in the MRD negative group compared with the MRD positive group by sequencing (TTP, median 80 vs. 31 months, p<0.0001; and OS, median not reached vs. 80 months p=0.02) (Figures 1A and 1B). All patients in VGPR, with the exception of 4 cases, were MRD positive by sequencing (92%). Three of the 4 cases with MRD negative and positive immunofixation became immunofixation negative after maintenance treatment. When restricting the analysis to the 61 patients that were in conventional CR (negative immunofixation), 35 of 60 patients were positive by sequencing at MRD levels at 10-5 and higher and 26 were MRD negative. There was a significantly improved TTP in the MRD negative group compared with the MRD positive group (median 131 vs. 35 months, p=0.001) (Figure 1C).
A high correlation between MFC and sequencing MRD results was observed (r2=0.87); as well as between ASO-PCR of Ig genes and sequencing (r2=0.89). Discordant results between FCM and sequencing (SEQ) included: 14 cases who were positive by SEQ and negative by FCM, and 5 cases negative by SEQ but positive by FCM; while the remaining cases were concordant: 63 cases were positive by both methods and 16 were negative by both methods. Correlation between ASO-PCR and SEQ was possible in 45 patients: 10 were discordant (9PCR-SEQ+ and 1 PCR+SEQ-) while 35 were concordant (20 PCR+SEQ+, 15 PCR-SEQ-).
The sequencing applicability for MRD studies in MM is 91%, which is essentially equivalent with that of MFC (96% Rawstron AC et al, JCO 2013). There is a high correlation between MRD levels by sequencing, ASO-PCR and MFC. Based on these results, MRD assessment by sequencing is a useful method for patient risk stratification and can be used to determine molecular CR in MM.
Pepin:Sequenta, Inc.: Employment, Equity Ownership. Weng:Sequenta, Inc.: Employment, Equity Ownership. Faham:Sequenta, Inc.: Employment, Equity Ownership, Membership on an entity’s Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal