Abstract
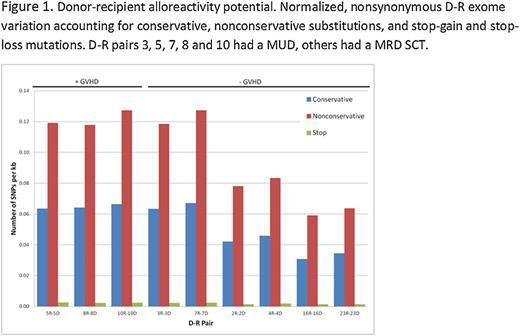
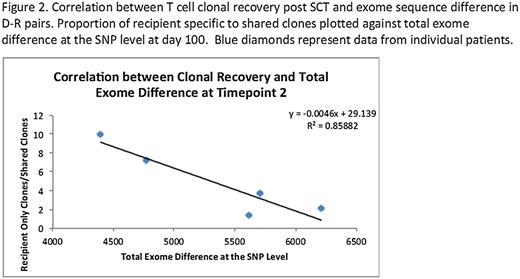
Graft versus host disease (GVHD) continues to afflict allogeneic hematopoietic stem cell transplant (SCT) recipients despite HLA matching at the molecular level. This is largely a consequence of minor histocompatibility antigen (mHA) variation between the donors and recipients resulting in alloreactivity. To determine the extent of potential antigenic variation at a molecular level, whole exome sequencing (WES) was performed on DNA samples from nine HLA-matched donor-recipient (D-R) pairs to catalogue the sequence variation. TruSeq exome enriched libraries were prepared using Illumina HiSeq 2000 platform. All exomes had at least 28X coverage; single nucleotide polymorphism (SNP) calling was performed using Genome Analysis Toolkit v1.6. Donor and recipient samples were compared with each other and reference human genome using ANNOVAR. SNP between donors and recipients were coded as either synonymous (S) or nonsynonymous (NS); and either NS conservative (NSc), or non-conservative (NSnc). Further, SNPs were analyzed with respect to graft versus host (GVH) or host vs. graft (HVG) vector. First, the recipient exomes were compared with the actual HLA matched and ‘simulated’ non-HLA matched donors; in this analysis, the major histocompatibility region demonstrated a 4-10 fold difference between the actual and simulated D-R pairs, unlike the whole exome, where similar sequence variation was observed between individuals regardless of HLA matching. For the actual D-R pairs the mean difference across the exome was 13,423 SNP, of which an average 6,445 were NS. Normalized for the number of nucleotide positions sequenced in the whole exome, the S- and NS-SNP scores were approximately equal, and NSnc-SNP were consistently > NSc-SNP (Figure 1). Further, in unrelated donor (URD) SCT, difference scores were approximately twice as large as in matched related donor recipients. GVHD was seen in 3/5 URD SCT with an alloreactivity score >0.1 NSnc-SNP/kilobase pair (Figure 1). Importantly the SNPs were equally distributed in GVH or HVG direction across the whole exome and the MHC locus in HLA matched pairs. When the location of the SNP was mapped onto the individual chromosomes, these were distributed across the entire genome with a small number of genes demonstrating frequent variation and others with a diminishing frequency. T cell receptor b sequencing was performed to determine T cell repertoire in donors, and recipients following SCT. Comparison of exome sequence variation between donor-recipient pairs and the proportion of unique to shared T cell clones and their frequency in recipients, suggested that post-transplant T cell repertoire reconstitution in the recipient may be influenced by exome sequence variation (Figure 2). In conclusion, WES reveals extensive nucleotide sequence variation in the exomes of HLA-matched donors and recipients indicating a large potential for alloreactivity in stem cell transplantation. By directly measuring this alloreactivity potential, in the future whole exome sequencing may aid in determining GVHD risk in HLA matched and mismatched D-R pairs.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal