Key Points
Transcription factors Batf3, Id2, and Nfil3 are not essential for induced CD8α+ DC generation.
Induced CD8α+ DCs can cross-present cellular antigens.
Abstract
Antiviral immunity and cross-presentation is mediated constitutively through CD8α+ and CD103+ DCs. Development of these DC subsets is thought to require the transcription factors Irf8, Id2, Nfil3, and Batf3, although how this network is regulated is poorly defined. We addressed the nature of the differentiation blocks observed in the absence of these factors and found that although all 4 factors are required for CD103+ DC development, only Irf8 is essential for CD8α+ DCs. CD8α+ DCs emerged in the absence of Id2, Nfil3 and Batf3 in short-term bone marrow reconstitution. These “induced” CD8α+ DCs exhibit several hallmarks of classic CD8α+ DCs including the expression of CD24, Tlr3, Xcr1, Clec9A, and the capacity to cross-present soluble, cell-associated antigens and viral antigens even in the absence of Batf3. Collectively, these results uncover a previously undescribed pathway by which CD8α+ DCs emerge independent of Id2, Nfil3, and Batf3, but dependent on Irf8.
Introduction
CD103+ and CD8α+ dendritic cells (DCs) are important for cross-presentation of antigens and for the induction of effector CD8+ T-cell responses against pathogens. Differentiation of these 2 subsets is regulated by a common set of transcription factors including interferon regulatory factor 8 (Irf8), nuclear factor interleukin 3-regulated (Nfil3 or E4BP4) and Id2 (inhibitor of DNA binding 2).1-4 Both populations also express the transcription factor Batf3 (basic leucine zipper transcriptional factor ATF-like 3, also known as Jun dimerization protein p21SNFT)2,5 but display differential dependence on Batf3 for differentiation.6,7 In initial studies, loss of Batf3 was thought to be critical for the development of CD8α+ DCs which provided an elegant explanation for the inability of Batf3−/− mice (generated on a B6.129 background) to cross-present cell-associated or viral antigens.5 Subsequently, these mice were discovered to also lack CD103+ DCs.2 This allowed confirmation of the hypothesis that CD103+ DCs from the lung were important for capture and transport of antigen to the draining lymph node (LN) where an immune response is initiated after inflammation or infection.8-10 Despite this, it has been observed that at steady-state in mice of a C57BL/6 genetic background, a population of DCs that express low levels of CD8α persist in spleen and LNs, and CD8α+ DC precursors can be identified in vitro in response to Flt3L stimulation.6,7 Furthermore, CD103 expression could be induced in vitro on DCs after addition of GM-CSF even in the absence of Batf3.6,7 Collectively, these findings raise major questions concerning the exact role of Batf3−/− in DC differentiation and function.
We now report that Batf3 intrinsically controls the development of CD103+Epcam+ DCs and the ability to cross-present cell-associated, but not soluble antigens, at steady-state. Soluble and viral antigens can be presented even in the absence of Batf3. After short-term bone marrow reconstitution, DCs derived from Batf3−/− bone marrow exhibited many hallmarks of wild-type CD8α+ DCs, although they failed to persist in hosts long-term. This effect was not limited to Batf3-deficient bone marrow but was also observed in the absence of Id2 or Nfil3. However, in the absence of Irf8, CD8α-expressing DCs did not emerge. These results establish that CD8α+-equivalent DCs found immediately after transplantation can bypass cues provided by the Id2-Nfil3-Batf3 pathway, but Irf8 was an essential determinant for their differentiation.
Methods
Mice
Id2gfp/gfp,7 Id2gfp/gfp × Batf3−/− (backcrossed > 10 generations to C57BL/6 mice; homozygous Id2gfp/gfpBatf3−/−),5 Id2gfp/gfp × Nfil3 (Id2gfp/gfpNfil3),11 Id2gfp/gfp × Irf8−/−(Id2gfp/gfpIrf8−/−), Ccr2-DTR-CFP,12 Cx3cr1-GFP,13 Myd88−/−,14 B6.CH-2bm−1 (bm1), Bm1.ActmOVA,15 Csf2−/− (GM-CSF KO),16 OT-I,17 OT-II,18 and B6.SJL-Ptprca (PTPrca, H-2b, CD45.1+) were used at 6 to 8 weeks. Id2fl/fl × CD11cCreT/+ (Id2fl/flCD11cCreT/+) were generated as described in supplemental Figure 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). All experiments were performed in accordance with institute guidelines and the Walter and Eliza Hall Institute of Medical Research Animal Ethics Committee.
Isolation of DCs from spleen and LNs
Spleens or LNs were chopped and then were digested for 20 minutes at room temperature with collagenase and DNase then treated for 5 minutes with EDTA to disrupt T cell–DC complexes.19,20 Light-density cells were isolated from cell suspensions using Nycodenz density (spleen 1.076 g/cm3 and LN 1.082 g/cm3) and were centrifuged at 1700g for 10 minutes at 4°C. The light-density cells were collected, counted, washed, and then depleted of non-DC lineages before staining for analysis.20
Generation of bone marrow chimeric mice
Bone marrow chimeras were generated from lethally irradiated (2 × 5.5 Gy, 3 hours apart) Csf2−/− or wild-type mice (Ly5.1 or Ly5.2) reconstituted with wild-type Id2gfp, Id2gfp/gfpBatf3−/−, Id2gfp/gfpNfil3, Id2gfp/gfpIrf8−/−, Id2fl/flCD11cCreT/+, Ccr2-DTR-CFP, Cx3cr1-GFP, or Myd88−/− bone marrow. Mixed bone marrow chimeric mice were generated by reconstituting recipients with a 1:1 ratio of congenically marked wild-type (Batf3+/+) and Batf3−/− bone marrow. For analyses of graft-versus-host disease (GVHD), Bm1.ActmOVA recipients were reconstituted with either Id2gfp or Id2gfp/gfpBatf3−/− bone marrow and 2 × 103 CD3+ T cells from Ly5.1+ mice.21 Chimeric mice were used for analysis 14 days up to 8 to 10 weeks after reconstitution.
Bone marrow cultures
Antigens and in vitro antigen presentation
Soluble OVA and OVA-coated spleen was prepared as previously described.7,23 Two to 2.5 × 104 purified DCs from the different subsets were washed and resuspended in 200 mL complete RPMI 1640 medium containing 2 × 105 OVA-coated irradiated bm1 splenocytes and 1 × 105 CFSE or CellTracker Violet (CTV)–labeled OT-I or OT-II T cells. After 60 hours in culture, T cells were stained for CD8α or CD4 and proliferation was analyzed by flow cytometry as previously described.24
Viral infections
Mice were injected subcutaneously with HSV-1 KOS strain (4 × 105 pfu) in each hind foot or 4 × 104 pfu for intravenous infection.25
Flow cytometric staining and cell sorting
DCs26,27 were stained with various combinations of mAbs to CD11c (HL3), CD45RA (14.8), CD11b (M1/70), CD24 (M1/69), CD4 (GK1.5), CD8α (53-6.7), signal regulatory protein (SIRP)–α (p84), CD103 (M290), CD24 (M1/69), Clec9A,28 MHC II (M5/114), and Ly5.1 (A20-1.1). Cells not of the DC lineage and dead cells were excluded by staining for CD19 (ID3), NK1.1 (NKR.PIC), CD3e (17A2), and propidium iodide (PI). Cell sorting and analysis were performed on a FACSCanto II or FACSAria (BD Instruments).
Quantitative RT-PCR
Total RNA was prepared from purified DC subsets using an RNeasy mini kit (QIAGEN). cDNA was synthesized from total RNA with OligoDT and thermoscript reverse transcriptase (Invitrogen). Real-time PCR was performed using the SensiMix SYBR no-Rox kit (Bioline). Analyses were done in triplicate and mean normalized expression was calculated with the Q-Gene application with Hprt as the reference gene.29 Primer sequences were Xcr1: forward 5′-CTCAGCCTTGTGGGTAACAGC-3′, reverse 5′-ACAGGCAGTAGACAGGAGAAC-3′30 ; Tlr3: forward 5′-AAAAACTCAGCGGCCGGAATG-3′, reverse 5′-AGTTACGAAGAGGGCGGAA-AGG-3′; Batf3: forward 5′-GCGCCCGGGAACCA3′, reverse 5′-AACCCGGTTTTTCTCTCTCCTT-3′; and Hprt: forward 5′-GGGGGCTATAAGTTCTTTGC-3′, reverse 5′-TCCAACACTTCGAGAGGTCC-3′.
Results
CD8α+ DCs develop in the absence of Batf3 but fail to survive long-term
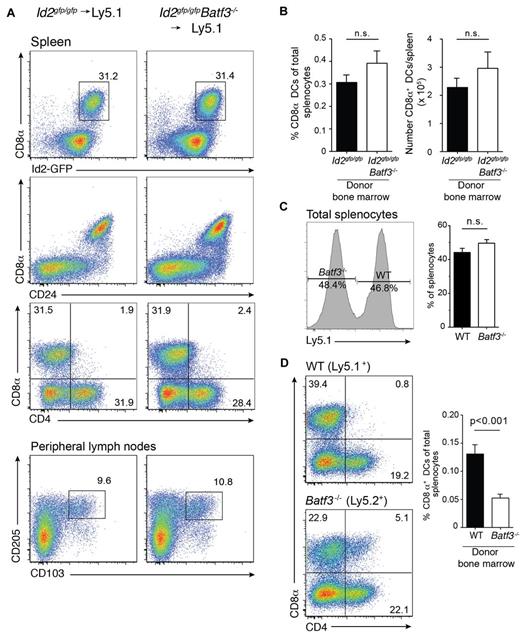
Batf3 is highly expressed in conventional DCs, with low to absent expression in other tissues of the body.5 Batf3−/− mice on a C57BL/6 background display a detectable, but significantly diminished, population of CD8α+ DCs with markedly reduced CD8α expression (Figure 1).6,7 These mice also preferentially lacked CD103+Epcam+ DCs (Figure 1). In vitro, a precursor DC population analogous to CD8α+ DCs could be identified as CD11c+Sirp-α− Id2+ cells,7 wheras CD103+ DCs did not develop in the absence of GM-CSF when Batf3−/− bone marrow progenitors were exposed to Flt3L.7 To determine whether the limited CD8α+ DC differentiation observed in vivo in the absence of Batf3 was an intrinsic property, or depended on extrinsic signals, bone marrow chimeric mice were generated in which lethally irradiated Ly5.1+ recipient mice were reconstituted with either Id2gfp/gfpBatf3−/− or Id2gfp/gfp bone marrow (Figure 1). Eight weeks after reconstitution, CD8α+ DCs in spleens and CD103+ DCs in LNs of Id2gfp/gfpBatf3−/− → Ly5.1 recipient mice were 2- to 4-fold reduced compared with recipient mice reconstituted with wild-type bone marrow (Figure 1). Taken together these results show that although Batf3 was initially thought to be essential for the development of CD8α+ and CD103+ DCs at steady state, under certain conditions CD8α+ DCs efficiently form in its absence.
Development of CD8α+ and CD103+ DCs in the presence of Batf3+/+ or Batf3−/− stromal cells. Analyses of purified CD11c+ DCs from intact Id2gfp/gfp, Id2gfp/gfpBatf3−/− mouse strains and bone marrow chimeric Id2gfp/gfp (Ly5.2+)→Ly5.1+ or Id2gfp/gfpBatf3−/−(Ly5.2+)→Ly5.1+ recipient mice 8 weeks after reconstitution. DCs were purified from spleens, peripheral LNs, and mesenteric LNs stained for various surface molecules and analyzed by flow cytometry. Numbers indicate the percentage of cells in each DC subset gated on live (PI−) donor (Ly5.1− or Ly5.2− cells) CD11c+CD45RA− conventional DCs. Data are representative of at least 3 separate experiments with similar results (n = 6-9 mice per group).
Development of CD8α+ and CD103+ DCs in the presence of Batf3+/+ or Batf3−/− stromal cells. Analyses of purified CD11c+ DCs from intact Id2gfp/gfp, Id2gfp/gfpBatf3−/− mouse strains and bone marrow chimeric Id2gfp/gfp (Ly5.2+)→Ly5.1+ or Id2gfp/gfpBatf3−/−(Ly5.2+)→Ly5.1+ recipient mice 8 weeks after reconstitution. DCs were purified from spleens, peripheral LNs, and mesenteric LNs stained for various surface molecules and analyzed by flow cytometry. Numbers indicate the percentage of cells in each DC subset gated on live (PI−) donor (Ly5.1− or Ly5.2− cells) CD11c+CD45RA− conventional DCs. Data are representative of at least 3 separate experiments with similar results (n = 6-9 mice per group).
We were able to detect CD8α+ DCs in chimeric mice at 8 weeks but their frequency was markedly reduced. In an earlier study,7 we found that the in vitro precursors of CD8α+ DCs that lack Batf3 die more rapidly than wild-type cells7 and proposed that the generation of CD8α+ DCs might be much greater at an earlier time-point as newly generated DCs repopulate organs and replace host DCs after only a few weeks.31,32 Therefore, we investigated whether expansion of CD8α+ DCs was initiated earlier during reconstitution of the hematopoietic compartment after transplantation (Figure 2A). Surprisingly, a major expansion of CD8α+ DCs was evident in the spleen at 3 weeks in both wild-type and Batf3-deficient chimeric mice with CD8α+ DCs representing a similar frequency and number of DCs found in spleen (Figure 2B). In addition, expansion of these CD8α+ DCs occurred when a mixture of bone marrow wild-type and Batf3−/− (1:1) was used to reconstitute recipient mice. It should be noted that although wild-type and Batf3−/− lymphocytes were equally represented in spleen (Figure 2C), generation of CD8α+ DCs proceeded less efficiently than their wild-type counterparts (Figure 2D) suggesting they may not compete as effectively for hematopoietic niches during reconstitution.
Expansion of CD8α+ DCs after short-term bone marrow reconstitution. Analyses of purified CD11c+ DCs from bone marrow chimeric Id2gfp/gfp (Ly5.2+)→Ly5.1+, Id2gfp/gfpBatf3−/− (Ly5.2+)→Ly5.1+ mice 21 days after reconstitution. DCs were purified from (A) spleen (top panels) and peripheral LNs (bottom panels) then stained for various surface molecules and analyzed by flow cytometry. Numbers indicate the percentage of cells in each DC subset gated on live (PI−) donor (Ly5.1− or Ly5.2− cells) CD11c+CD45RA− DCs. (C) Proportion and total number of CD8α+ DCs recovered from spleens of chimeric mice as in panel A. Data are representative of at least 3 separate experiments with similar results (n = 6 mice per group). (D) Analysis of splenocytes (left panel) and DCs (right panel) purified from spleens of Ly5.1+:Batf3−/− (Ly5.2+)→Ly5.1/5.2+ F1 mixed bone marrow chimeras and analyzed by flow cytometry. Numbers indicate the percentage of cells in each DC subset gated on live (PI−) donor (Ly5.1+ or Ly5.2+ cells) bone marrow. Data are representative of at least 2 separate experiments with similar results (n = 5-6 mice per group).
Expansion of CD8α+ DCs after short-term bone marrow reconstitution. Analyses of purified CD11c+ DCs from bone marrow chimeric Id2gfp/gfp (Ly5.2+)→Ly5.1+, Id2gfp/gfpBatf3−/− (Ly5.2+)→Ly5.1+ mice 21 days after reconstitution. DCs were purified from (A) spleen (top panels) and peripheral LNs (bottom panels) then stained for various surface molecules and analyzed by flow cytometry. Numbers indicate the percentage of cells in each DC subset gated on live (PI−) donor (Ly5.1− or Ly5.2− cells) CD11c+CD45RA− DCs. (C) Proportion and total number of CD8α+ DCs recovered from spleens of chimeric mice as in panel A. Data are representative of at least 3 separate experiments with similar results (n = 6 mice per group). (D) Analysis of splenocytes (left panel) and DCs (right panel) purified from spleens of Ly5.1+:Batf3−/− (Ly5.2+)→Ly5.1/5.2+ F1 mixed bone marrow chimeras and analyzed by flow cytometry. Numbers indicate the percentage of cells in each DC subset gated on live (PI−) donor (Ly5.1+ or Ly5.2+ cells) bone marrow. Data are representative of at least 2 separate experiments with similar results (n = 5-6 mice per group).
Although loss of Batf3 has been reported to influence cross-presentation at steady-state and to ablate viral antigen presentation, these settings used cell-associated antigens, or pathogens that required the CD103+Epcam+ DCs to reach LN DCs for initiation of an immune response.5,9 This population is largely lacking in Batf3−/− mice (Figure 1) and thus the previous experimental approaches do not directly test viral antigen presentation in vivo. Given these circumstances, we asked whether presentation of antigens, which may occur by direct presentation, cross-presentation, or “cross-dressing” of soluble and viral antigens was also ablated in the absence of Batf3.
Batf3-deficiency does not affect cross-presentation of soluble antigens at steady-state.
Initially, we tested the capacity of CD8α+-equivalent DCs7 isolated from Flt3L/GM-CSF bone marrow cultures of wild-type (Ly5.2→Ly5.1) or Batf3−/−→Ly5.1 chimeric mice to cross-present soluble antigens. Coculture of DC subsets purified from day 8 cultures with OVA and either OT-I or OT-II (not shown) T cells revealed that both wild-type and Batf3−/− CD8α+-equivalent in vitro–derived DCs could efficiently cross-present soluble antigens (Figure 3A). Similar results were observed in cultures from intact mice highlighting that at least in this setting, the cross-presentation pathway for soluble antigens remained intact.
In vitro and in vivo cross-presentation of cell-associated, soluble, and viral antigens in Batf3-deficient mice. (A) Ly5.2+CD45RA− CD11c+Sirpα+ (top panels), CD11c+Sirpα−CD103− (CD8α+ equivalent; middle panels) and CD11c+Sirpα−CD103+ DCs (bottom panels) were purified from bone marrow cultures of Id2gfp/gfp(Ly5.2+)→Ly5.1 or Id2gfp/gfpBatf3−/−(Ly5.2+)→Ly5.1 chimeric mice that had been allowed to reconstitute for 10 weeks. Bone marrow was stimulated with Flt3L for 8 days with the addition of GM-CSF on day 6. FACS purified populations of DCs were cocultured with soluble OVA together with either CD8+ OT-I cells labeled with CellTracker Violet (CTV). Numbers represent the percentage of cells that have proliferated in response to exogenous antigen. Data are representative of bone marrow cells derived from 2 separate cohorts of Id2gfp/gfp(Ly5.2+)→Ly5.1 or Id2gfp/gfpBatf3−/−(Ly5.2+)→Ly5.1 chimeric mice and each DC subset analyses was performed in duplicate or triplicate within each experiment. (B) In vivo cross-presentation of soluble OVA (top panels). OVA-coated irradiated bm-1 splenocytes or soluble OVA was injected into recipient wild-type or Batf3−/− mice containing 1.5 × 106 CTV-labeled Ly5.1+ OT-I cells. After 3 days, the spleen was isolated and analyzed for proliferation by loss of CTV from Ly5.1+Vα2+CD8+ T cells. (C) Cross-presentation of cell-associated OVA in GVHD. Bm1.ActmOVA mice were irradiated and transplanted with 5 × 106 BM from either Id2-GFP or Id2 × Batf3−/− bone marrow, and 2 × 103 CD3+ T cells from Ly5.1 congenic mice. CSFE-labeled Ly5.1+ OT-I cells were adoptively transferred into mice on day 10 after transplantation and proliferation in the OVA-specific Vα2+ CD8+ T-cell population analyzed after 3 days. Data show representative proliferation profiles (left panels) and the mean number ± SD of Kb-OVA Vα2+ CD8+ T cells recovered per spleen (n = 8). NS, not significant. (D) Responses to HSV-1 virus infection. 1.5 × 106 CTV labeled Ly5.1+ HSV-1–specific (glycoprotein B, gB) CD8α+ T cells were adoptively transferred into wild-type or Batf3−/− mice 1 day before subcutaneous foot pad inoculation with HSV-1. After 3 days, the spleen was isolated and analyzed for proliferation by loss of CTV from Ly5.1+Vα2+CD8+ T cells. Data are representative of 3 experiments with 2 to 4 mice in each group for each experiment.
In vitro and in vivo cross-presentation of cell-associated, soluble, and viral antigens in Batf3-deficient mice. (A) Ly5.2+CD45RA− CD11c+Sirpα+ (top panels), CD11c+Sirpα−CD103− (CD8α+ equivalent; middle panels) and CD11c+Sirpα−CD103+ DCs (bottom panels) were purified from bone marrow cultures of Id2gfp/gfp(Ly5.2+)→Ly5.1 or Id2gfp/gfpBatf3−/−(Ly5.2+)→Ly5.1 chimeric mice that had been allowed to reconstitute for 10 weeks. Bone marrow was stimulated with Flt3L for 8 days with the addition of GM-CSF on day 6. FACS purified populations of DCs were cocultured with soluble OVA together with either CD8+ OT-I cells labeled with CellTracker Violet (CTV). Numbers represent the percentage of cells that have proliferated in response to exogenous antigen. Data are representative of bone marrow cells derived from 2 separate cohorts of Id2gfp/gfp(Ly5.2+)→Ly5.1 or Id2gfp/gfpBatf3−/−(Ly5.2+)→Ly5.1 chimeric mice and each DC subset analyses was performed in duplicate or triplicate within each experiment. (B) In vivo cross-presentation of soluble OVA (top panels). OVA-coated irradiated bm-1 splenocytes or soluble OVA was injected into recipient wild-type or Batf3−/− mice containing 1.5 × 106 CTV-labeled Ly5.1+ OT-I cells. After 3 days, the spleen was isolated and analyzed for proliferation by loss of CTV from Ly5.1+Vα2+CD8+ T cells. (C) Cross-presentation of cell-associated OVA in GVHD. Bm1.ActmOVA mice were irradiated and transplanted with 5 × 106 BM from either Id2-GFP or Id2 × Batf3−/− bone marrow, and 2 × 103 CD3+ T cells from Ly5.1 congenic mice. CSFE-labeled Ly5.1+ OT-I cells were adoptively transferred into mice on day 10 after transplantation and proliferation in the OVA-specific Vα2+ CD8+ T-cell population analyzed after 3 days. Data show representative proliferation profiles (left panels) and the mean number ± SD of Kb-OVA Vα2+ CD8+ T cells recovered per spleen (n = 8). NS, not significant. (D) Responses to HSV-1 virus infection. 1.5 × 106 CTV labeled Ly5.1+ HSV-1–specific (glycoprotein B, gB) CD8α+ T cells were adoptively transferred into wild-type or Batf3−/− mice 1 day before subcutaneous foot pad inoculation with HSV-1. After 3 days, the spleen was isolated and analyzed for proliferation by loss of CTV from Ly5.1+Vα2+CD8+ T cells. Data are representative of 3 experiments with 2 to 4 mice in each group for each experiment.
Batf3-deficiency affects cross-presentation of cell-associated at steady-state.
It was surprising that in vitro, Batf3−/− CD8α+-equivalent DCs could cross-present soluble antigens although cross-presentation of cell-associated antigens appears to be impaired.5-7 This suggested that, in addition to being selectively required for the development of CD103+Epcam+ DCs, Batf3-deficiency mechanistically separated the pathways required for cross-presentation of cell-associated and soluble antigens. To investigate whether this later pathway persisted in vivo, we tested intact mice for their ability to cross-present the different forms of antigen, namely cell-associated and soluble OVA. Analyses of intact wild-type and Batf3−/− mice confirmed that soluble, but not cell-associated, OVA was efficiently cross-presented in the absence of Batf3 (Figure 3B).
Batf3 is not required for cross-presentation of cell-associated antigens during GVHD
To extend this analysis, we analyzed whether cross-presentation of cell-associated antigens remained intact in in the setting of the intense inflammation that follows allogeneic bone marrow transplantation (BMT) and GVHD (Figure 3C).21 In this model of allogeneic BMT, ovalbumin is ubiquitously expressed on host tissue (driven of the β-actin promoter, Act-mOVA15 ), but host cells (H2-Kbm1) lack the appropriate MHC class I molecule to present OVA-derived peptides to OT-I CD8+ T cells. Thus, any antigen presentation to the OT-I T cell must occur via donor antigen presenting cells bearing C57BL/6 MHC class I. Batf3+/+ and Batf3−/− CD8α+ DCs developed in the Act-mOVA hosts similar to non-antigen expressing hosts (ie, Ly5.1+ mice). In addition, OT-I cells adoptively transferred into mice transplanted with wild-type or Batf3−/− bone marrow proliferated equally well indicating that Batf3 is not required for either the development of CD8α+ DCs or cross-presentation during experimental GVHD.
Batf3 is not required for presentation of HSV-1 viral antigens in vivo
We then investigated whether induction of antiviral CD8+ T-cell responses could occur in Batf3−/− mice. We have shown previously that although initiation of a CD8+ T-cell response depends on trafficking of antigen via migratory CD103+ DCs and hand-over to CD8α+ DCs, subcutaneous infection with HSV-1 bypasses the requirement for the CD103+Epcam+ DCs to traffic to the LN, and exposes LN resident DCs directly to the virus.33-36 Circumventing the necessity for DC trafficking showed that LN DCs could efficiently activate HSV-specific glycoprotein B (gB) CD8+ T cells even in the absence of Batf3 (Figure 3D; proliferating gBT-1 CD8+ T cells/popliteal LN, mean ± SD: WT 2.11 × 104 ± 1.63 × 103; Batf3−/− 2.21 × 104 ± 1.01 × 104). This might be accounted for by the capacity for CD8α+ DCs to be expanded after viral infection (supplemental Figure 2). Thus, in contrast to earlier reports, viral antigen presentation and the induction of CD8α+ DCs is possible in the absence of Batf3.
To better understand how the emergence of these induced CD8α+ DCs might be regulated, we analyzed the role of the cytokines TSLP, GM-CSF, Flt-3L, or irradiation alone in this process (supplemental Figure 3A-C, and data not shown). Amplification of CD8α+ DCs could not be achieved in Batf3−/− mice by the delivery of TSLP via hydrodynamic injection or by the provision of Flt3L that normally greatly expands the CD8α+ DC population in wild-type mice (data not shown). As the CD8α+ DC expansion was observed in irradiated mice, but not at steady-state, we reasoned that irradiation alone might be sufficient to induce this effect. To examine this, wild-type and Batf3−/− mice were sublethally irradiated and the DC compartment analyzed after 20 days. In this setting, irradiation alone was not sufficient to induce the expansion of CD8α+ DCs (supplemental Figure 3) indicating that DC reconstitution from the transplanted bone marrow was critical for this process. Induced CD8α+ DCs were also observed after reconstitution of GM-CSF–deficient mice with Batf3−/− bone marrow, ruling out an essential role for recipient GM-CSF in this process (supplemental Figure 3C). Similarly, reconstitution of recipients with bone marrow lacking expression of the universal adapter protein Myd88, which is required for signaling by most toll-like receptors, or bone marrow lacking expression of the inflammatory cytokine interferon-γ not impair the development of CD8α+ DCs after transplantation (supplemental Figure 3D and E, respectively).
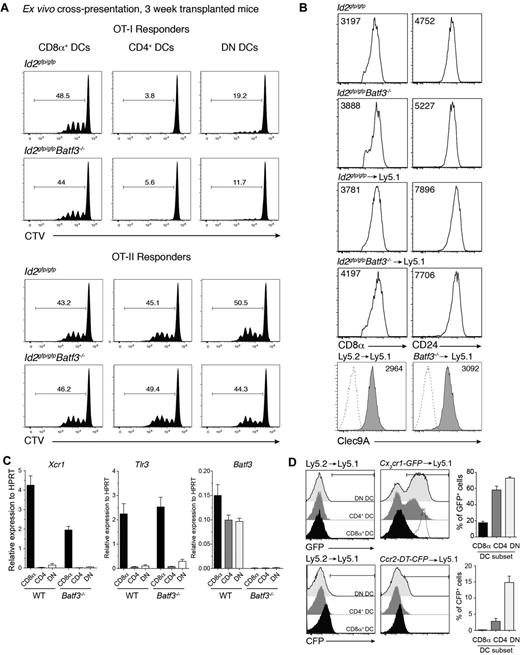
The expansion of a population of DCs that expressed CD8α in the absence of Batf3 was surprising particularly given that Batf3 was originally described as essential for the development of CD8α+ DCs.5 Therefore, we further investigated whether this population exhibited other hallmarks of the CD8α+ DC population that occurs at steady-state. CD8α-expressing DCs were purified from bone marrow chimeric mice then analyzed for their capacity to cross-present cell-associated antigens to OVA-specific CD8+ and CD4+ T cells. Strikingly, CD8α+ DCs isolated from mice that received bone marrow that lacked Batf3 expression were equally efficient as wild-type CD8α+ DCs in presenting OVA to both T-cell subsets (Figure 4A). These Batf3−/− CD8α+ DCs expressed key molecules including CD8α, CD24, Clec9A, Id2-GFP, Xcr1, and Tlr3 typically expressed by classic CD8α+ DCs but not monocytes37 and were indistinguishable (except for their lack of Batf3) from those isolated from mice reconstituted with wild-type bone marrow (Figures 1 and 4B-C). Furthermore, these CD8α+ DCs did not express chemokine receptors Ccr2 or Cx3cr1 (Figure 4D) found on monocyte subsets.38,39
Early induced CD8α+ DCs exhibit hallmarks of classic CD8α+ DCs. (A) Ly5.2+CD11c+CD45RA− CD8α+, CD4+ and CD8α−CD4− DN DCs derived from Id2gfp/gfp (Ly5.2+)→Ly5.1 or Id2gfp/gfpBatf3−/− (Ly5.2+)→Ly5.1 chimeric mice 3 weeks after transplantation of bone marrow were purified by FACS sorting from spleen. DCs (1.25 × 104) were cocultured with OVA-coated bm1 splenocytes and 5 × 103 CTV CD8+ OT-I or CD4+ OT-II T cells. After 60 hours, the level of proliferation induced in cultures were analyzed by staining cells for Vα2+ within the CD4+ and CD8+ populations. Data are representative profiles of 2 experiments with similar results. (B) Expression of CD8α, CD24, and Clec9A surface molecules in splenic CD8α+ DCs. Ly5.2+ CD11c+ splenic DCs from bone marrow chimeric mice as described in panel A. Numbers indicate the mean fluorescence intensity of each marker within the PI−CD45−CD11c+CD8α+ DC subset. (C) Quantitative PCR analyses of Xcr1, Tlr3, and Batf3 expression by CD8α+ DCs 3 weeks after bone marrow transplantation. FACS purified CD8α+ DCs isolated from Id2gfp/gfp (Ly5.2+) or Id2gfp/gfpBatf3−/− (Ly5.2+)→Ly5.1 recipients and analyzed for expression of Tlr3. (D) Analysis of CX3CR1 and CCR2 expression on splenic DC subset 3 weeks after reconstitution of Ly5.1 recipients with Ly5.1 (WT), Cx3cr1-GFP, or Ccr2-DTR-CFP bone marrow. Dotted line shows expression on Ly6Chigh monocytes. (A-D) Data are representative of at least 2 experiments for each condition and error bars represent mean ± SD.
Early induced CD8α+ DCs exhibit hallmarks of classic CD8α+ DCs. (A) Ly5.2+CD11c+CD45RA− CD8α+, CD4+ and CD8α−CD4− DN DCs derived from Id2gfp/gfp (Ly5.2+)→Ly5.1 or Id2gfp/gfpBatf3−/− (Ly5.2+)→Ly5.1 chimeric mice 3 weeks after transplantation of bone marrow were purified by FACS sorting from spleen. DCs (1.25 × 104) were cocultured with OVA-coated bm1 splenocytes and 5 × 103 CTV CD8+ OT-I or CD4+ OT-II T cells. After 60 hours, the level of proliferation induced in cultures were analyzed by staining cells for Vα2+ within the CD4+ and CD8+ populations. Data are representative profiles of 2 experiments with similar results. (B) Expression of CD8α, CD24, and Clec9A surface molecules in splenic CD8α+ DCs. Ly5.2+ CD11c+ splenic DCs from bone marrow chimeric mice as described in panel A. Numbers indicate the mean fluorescence intensity of each marker within the PI−CD45−CD11c+CD8α+ DC subset. (C) Quantitative PCR analyses of Xcr1, Tlr3, and Batf3 expression by CD8α+ DCs 3 weeks after bone marrow transplantation. FACS purified CD8α+ DCs isolated from Id2gfp/gfp (Ly5.2+) or Id2gfp/gfpBatf3−/− (Ly5.2+)→Ly5.1 recipients and analyzed for expression of Tlr3. (D) Analysis of CX3CR1 and CCR2 expression on splenic DC subset 3 weeks after reconstitution of Ly5.1 recipients with Ly5.1 (WT), Cx3cr1-GFP, or Ccr2-DTR-CFP bone marrow. Dotted line shows expression on Ly6Chigh monocytes. (A-D) Data are representative of at least 2 experiments for each condition and error bars represent mean ± SD.
Transcriptional regulators are major determinants of the differentiation program of DC subsets.1 Nevertheless, our data support the notion that at least in the case of Batf3, development of cells that express markers and functions representative of CD8α+ DCs can proceed in the absence of this transcriptional regulator and is thus not intrinsically dependent on Batf3. To determine whether the CD8α+ DCs that emerge after bone marrow transplantation occurs only in the absence of Batf3, we investigated the role other transcription factors such as Id2, Nfil3, and Irf8 conventionally required for the development of the CD8/CD103 DC lineage (Figure 5).1-3,5,7,40 Lethally irradiated Ly5.1 recipients were reconstituted with bone marrow lacking Irf8, Id2 (see supplemental Figure 2) or Nfil3. We observed that DCs isolated from naive Nfil3−/− mice also lacked CD103+ DCs, in addition to their loss of CD8α+ DCs, as was previously reported3 (data not shown). After 20 days, the reconstitution profiles of transplanted mice were examined by flow cytometry (Figure 5). Surprisingly, CD8α+ DCs arose in the absence of Id2 and Nfil3, but not Irf8, and Id2-GFP expression was maintained in the absence of Nfil3 (Figure 5).
Early CD8α+ DC expansion after transplantation depends on Irf8 and Nfil3 but not Id2. (A) Analysis of CD11c+CD45RA− DCs from spleens of bone marrow chimeric mice in which Id2gfp/gfp, Id2fl/flCD11cCreT/+, Id2gfp/gfpNfil3−/−, or Id2gfp/gfpIrf8−/−-deficient bone marrow was transplanted into lethally irradiated recipients (Ly5.1+) 15 to 20 days earlier. Cells were stained for various surface molecules and analyzed by flow cytometry. Data are representative of 2 to 3 separate experiments with 2 to 4 mice in each experiment with similar results (n = 6-8 mice per group). (B) Analysis of CD8α expression on DCs in the absence of Nfil3 or Id2. DCs were isolated from spleens of intact Id2gfpNfil3−/− or Id2CD11cCreT/+ mice. Data are representative of 2 experiments with similar results (n = 5 mice per genotype). (C) Proportion (top panel) and total number (bottom panel) of splenic CD8α+ DCs recovered from spleens of intact WT and KO mice or bone marrow chimeric (KO→Ly5.1) mice 3 weeks after transplantation. Data are pooled from 2 experiments with 5 mice per genotype; NS indicates not statistically significant.
Early CD8α+ DC expansion after transplantation depends on Irf8 and Nfil3 but not Id2. (A) Analysis of CD11c+CD45RA− DCs from spleens of bone marrow chimeric mice in which Id2gfp/gfp, Id2fl/flCD11cCreT/+, Id2gfp/gfpNfil3−/−, or Id2gfp/gfpIrf8−/−-deficient bone marrow was transplanted into lethally irradiated recipients (Ly5.1+) 15 to 20 days earlier. Cells were stained for various surface molecules and analyzed by flow cytometry. Data are representative of 2 to 3 separate experiments with 2 to 4 mice in each experiment with similar results (n = 6-8 mice per group). (B) Analysis of CD8α expression on DCs in the absence of Nfil3 or Id2. DCs were isolated from spleens of intact Id2gfpNfil3−/− or Id2CD11cCreT/+ mice. Data are representative of 2 experiments with similar results (n = 5 mice per genotype). (C) Proportion (top panel) and total number (bottom panel) of splenic CD8α+ DCs recovered from spleens of intact WT and KO mice or bone marrow chimeric (KO→Ly5.1) mice 3 weeks after transplantation. Data are pooled from 2 experiments with 5 mice per genotype; NS indicates not statistically significant.
Discussion
The studies reported here provide important insights into key differences in the control of CD8α+ DCs differentiation at steady-state and during transplantation. We show that DCs that exhibit the properties of CD8α+ DCs are rapidly expanded after transplantation but appear not intrinsically depend on several of the transcriptional regulators essential for CD8α+ DCs at steady-state. In addition, although Batf3 is essential for the development of splenic CD8α+ DCs, cross-presentation and presentation of viral antigens remains intact in vivo.
Cross-presentation of antigens in vivo has been attributed mainly to 2 subsets of DCs, namely the lymphoid-tissue resident CD8α+ DCs and the migratory CD103+CD11b− DCs. They play a fundamental role in immune responses to viruses.8,33,41,42 Intact Batf3−/− mice display a significant impairment in the ability to cross-present cell-associated antigens and viral antigens in distinct settings.5,9,43 Our data demonstrate that surprisingly, despite these initial observations, Batf3−/− mice retain the capacity to efficiently cross-present both soluble antigens and cell-associated antigens during GVHD. In addition, by targeting virus delivery to bypass the requirement for migratory DCs to deliver antigen to lymph node resident DCs, antigen presentation to CD8+ T cells appears normal. This might be attributable to the persistence of CD103+ DCs and precursors of CD8α+ DCs in the absence of Batf3.6,7,44 In unmanipulated Batf3−/− mice, CD8α+ DCs that express the marker Clec9A (also known as DNGRI45,46 ) are detectable and splenic precursors to CD8α+ (pre-CD8α+, CD8−CD24+MHCII+) DCs are expanded 3-fold44 implying that Batf3 either regulates the development of this subset46 or that Clec9A is a direct target of Batf3. An alternative proposal is that Batf3 may have specific effects on the cross-presenting pathway for cell-associated antigens accounting for the lack of cross-presentation observed in in vitro cultures. The latter seems unlikely given that after transplantation of Batf3-deficient cells, CD8α+ DCs (that also express Clec9A) have the capacity to cross-present cell-associated antigens equivalent to wild-type CD8α+ DCs.
The successive steps that lead to commitment of DC subsets in other settings such as during transplantation and pathogen infection, and the relationship between different DC subsets, remains poorly understood. Id2, Nfil3, Batf3, and Irf8 are all key regulators of the differentiation of steady-state CD8α+ and CD103+ DCs.1,4,47,48 Batf3 appears to be required only for the final differentiation step of DC development,6,7 whereas the maintenance of Id2-GFP expression in the absence of Nfil3 indicates that Id2 lies upstream of Nfil3 in driving DC differentiation. We show that after bone marrow transplantation, the emergence of CD8α+ DCs is not completely dependent on all elements of this transcriptional network, although notably induced DCs exhibit phenotypic features, and an antigen presenting program, similar to classic CD8α+ DCs.4 Supporting these findings, it has been demonstrated that in human DCs although inhibition of Batf3 prevented the development of the CD8α+CD11b− equivalent subset in vitro, Batf3 silencing was insufficient to impair the development of Clec9A+ DCs in humanized mice46 highlighting the capacity of Clec9A+ DCs in man to bypass the requirement for Batf3. Building on these observations, our data strongly imply that the requirement for Id2 and Nfil3 may also be circumvented in the development of Clec9A+ DCs in humans, at least in vivo, providing an alternate pathway for the development of cross-presenting DCs where genetic components of the classic pathway are disrupted.49 Our findings suggest that the transcriptional regulators of CD8α+ DCs can be divided into 2 groups. One group consisting of Irf8 and PU.150,51 which are absolutely required for CD8α+ DC differentiation, whereas the second group consisting of Id2, Nfil3, and Batf3 are essential for most, but not all, CD8α+ DC differentiation in steady-state but are dispensable during DC reconstitution after bone marrow transplantation.
The CD8α+ DCs induced after transplantation exhibit many features of classic CD8α+ DCs including the expression of classic surface molecules, such as CD24 and Tlr3, in addition to the ability to cross-present cell-associated antigens. Preliminary analyses suggest that this early expansion of CD8α+ DCs with these properties after transplantation may also be important in protecting against GVHD (GTB and KPAM, unpublished). Future work will be required to address the mechanism by which Id2-Nfil3-Batf3–independent CD8α+ DCs are expanded after bone marrow transplantation. Our data does rule out a dominant role of the DC promoting cytokines GM-CSF, Flt3L, TSLP, Myd88, or IFN-γ or irradiation alone in the expansion of these CD8α+ DCs. In some settings, other members of the Batf family, particularly Batf2, may substitute for Batf3 in the generation of CD8α+ and CD103+ DCs.52 Whether Batf2 also compensates for loss of Id2 and Nfil3 is currently unclear. However, it seems probable that the Id2-Nfil3-Batf3–independent nature of the CD8α+ DCs that develop may represent an alternative pathway of development requiring a different gene expression program and may be akin to monocyte-derived inflammatory DCs, or illustrate a partial dependency of conventional CD8α+ DCs on these single factors as has been proposed for intestinal DCs.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank M. Camilleri for assistance with animal work, Dr Wu for advice and discussion, Drs Lahoud and Caminschi for the kind gift of the Clec9A antibody, and the WEHI Flow Cytometry facility for cell sorting.
This work is supported by the National Health and Medical Research Council (NHMRC) of Australia. K.A.M. is a NHMRC Training Fellow; K.P.A.M. is a Cancer Council Queensland Senior Research Fellow; and G.R.H. is a NHMRC Australia Fellow and Queensland Health Senior Clinical Fellow. G.T.B is supported by a Sylvia and Charles Viertel Foundation Fellowship; and S.L.N. is supported by an ARC Future Fellowship. This work was made possible through Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIIS.
Authorship
Contribution: C.S., J.T.J., K.A.M., G.R.H., K.P.A.M., S.L.N., and G.T.B. designed research, performed research, prepared an initial draft based on a systematic review of published literature, and discussed the draft; and H.J.M.B. generated the Nfil3−/− mice.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gabrielle Belz, Division of Molecular Immunology, Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Melbourne, Victoria 3052 Australia; e-mail: belz@wehi.edu.au.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal