To the editor:
Notch-derived signals are essential for specification of hematopoietic progenitors to T-cell lineage and for promotion of β-selection at the CD4–CD8– double-negative 3 (DN3) stage. However, these signals are not required for further thymocyte maturation.1 Accordingly, the expression of Notch1 and its target genes, including Ptcra (encoding pre–T-cell receptor alpha [TCRα]), markedly decreases in late DN3 and DN4 cells.2,3 Notch1 downregulation has been attributed to pre-TCR–induced Id3, which antagonizes the Notch1-positive regulator, E2A.3 Hes1, a Notch target gene, is negatively regulated by Ikaros in DN4 cells.4 However, there are 2 unresolved issues regarding attenuation of Notch signals: (1) although reduced in transcripts, surface expression of Notch1 protein remains equally high in DN3 and DN4 cells,5 suggesting that downregulation of Notch target genes requires other repressive mechanisms independent of Notch11 ; and (2) because pre-TCR signals are attenuated/terminated in DN4 cells due to Ptcra downregulation,2 sustained Notch1 repression may require additional factors.
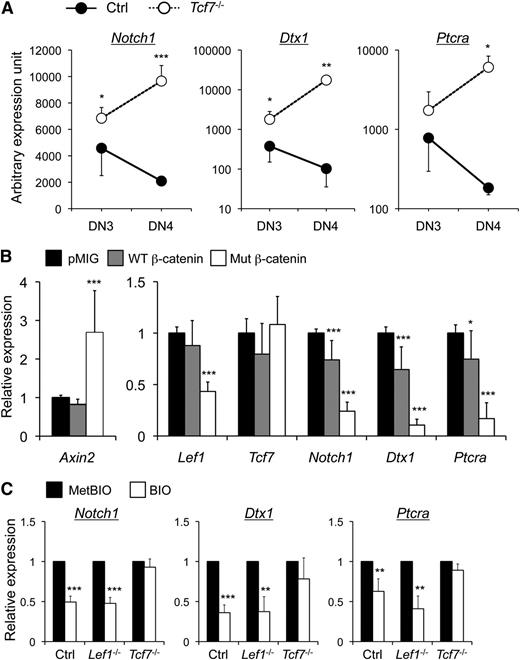
We recently demonstrated that T-cell factor 1 (TCF-1; encoded by Tcf7) suppressed thymic malignancy.6 The neoplastic cells exhibited increased expression of Notch1 and its target genes and accumulated somatic Notch1 mutations,6,7 suggesting possible negative regulation of Notch signaling by TCF-1. Analysis of premalignant Tcf7−/− thymocytes revealed that TCF-1 deficiency caused increased expression of Notch1 and its known targets, Dtx1 and Ptcra, in DN3 and more markedly in DN4 cells (Figure 1A). TCF-1 interacts with Wnt-modulated β-catenin cofactor to regulate gene expression. To substantiate negative regulation of Notch pathway by TCF-1, we infected primary DN thymocytes with retrovirus expressing WT or constitutively active mutant β-catenin.8 As expected, forced expression of mutant β-catenin induced Axin2 and repressed Lef1 expression, with little effect on Tcf7 itself in DN3 thymocytes (Figure 1B). Interestingly, the mutant β-catenin greatly diminished expression of Notch1, Dtx1, and Ptcra in DN3 and DN4 thymocytes (Figure 1B), indicating direct involvement of the TCF-1–β-catenin complex in Notch repression. To exclude the possibility that β-catenin acts through factors other than TCF-1, we stabilized β-catenin in Tcf7−/− DN3 cells using BIO, a GSK3β inhibitor. Compared with its inactive analog MetBIO, BIO stimulation diminished the expression of Notch1, Dtx1, and Ptcra in control DN3 thymocytes (Figure 1C). In contrast, this effect was abolished in Tcf7−/− DN3 thymocytes but relatively unaffected in LEF-1–deficient DN3 cells (Figure 1C).
TCF-1 and activated β-catenin negatively regulate Notch1 and its target genes in DN3 thymocytes. (A) Elevated expression of Notch1 and its target genes in TCF-1–deficient DN3 and DN4 thymocytes. Tcf7−/− mice were age 8 weeks or younger and without overt signs of thymic malignancy at the time of analysis. DN subsets were sorted from lineage-negative thymocytes from Tcf7−/− mice or littermate controls and assessed for gene expression. The relative expression level of individual genes was obtained by normalizing to the Hprt1 housekeeping gene. Data are means ± standard deviation from 1 of 3 experiments with similar results (n ≥ 3 in each experiment). (B) Activated β-catenin represses the expression of Notch1 and its targets in DN3 thymocytes. Lineage-negative DN thymocytes were cultured on OP9-DL1 stromal cells10 overnight in the presence of interleukin-7 (5 ng/mL) and then infected with empty retroviral vector pMIG or that expressing wild-type (WT) or mutant β-catenin. The mutant β-catenin has internal deletions of its N-terminal Ser/Thr phosphorylation sites and is therefore constitutively active.8 Twenty-four hours later, the GFP+ DN3 thymocytes were sorted and analyzed for expression of indicated genes. After normalization to Hprt1, the expression of each gene in pMIG-infected cells was arbitrarily set to 1, and its relative expression in the presence of WT or mutant β-catenin was then calculated. Data were pooled from at least 3 independent experiments (n ≥ 7). Similar data were obtained with DN4 cells (not shown). (C) β-catenin–mediated Notch repression depends on TCF-1. DN3 thymocytes were sorted from control mice, Vav1-Cre Lef1−/−6 or Tcf7−/−, cultured in the presence of 5 µM MetBIO or BIO for 6 hours, and then harvested for gene expression analysis. After normalizing to Hprt1, the expression of each gene in MetBIO-treated cells was arbitrarily set to 1, and its relative expression in BIO-treated samples was then calculated. Data are pooled from 2 independent experiments (n ≥ 3). *, P < .05; **, P < .01; ***, P < .001 by Student t test. Note that although multiple TCF/LEF binding motifs were found within “–30 kb ∼ +10 kb” of transcription initiation sites of the Notch1, Dtx1, and Ptcra genes, we did not find enriched binding of TCF-1 to these motifs in DN3 thymocytes. Further studies are necessary to determine if repression of Notch1 and its targets by TCF-1 is mediated by direct regulation via more distal TCF/LEF motifs or by indirect mechanisms.
TCF-1 and activated β-catenin negatively regulate Notch1 and its target genes in DN3 thymocytes. (A) Elevated expression of Notch1 and its target genes in TCF-1–deficient DN3 and DN4 thymocytes. Tcf7−/− mice were age 8 weeks or younger and without overt signs of thymic malignancy at the time of analysis. DN subsets were sorted from lineage-negative thymocytes from Tcf7−/− mice or littermate controls and assessed for gene expression. The relative expression level of individual genes was obtained by normalizing to the Hprt1 housekeeping gene. Data are means ± standard deviation from 1 of 3 experiments with similar results (n ≥ 3 in each experiment). (B) Activated β-catenin represses the expression of Notch1 and its targets in DN3 thymocytes. Lineage-negative DN thymocytes were cultured on OP9-DL1 stromal cells10 overnight in the presence of interleukin-7 (5 ng/mL) and then infected with empty retroviral vector pMIG or that expressing wild-type (WT) or mutant β-catenin. The mutant β-catenin has internal deletions of its N-terminal Ser/Thr phosphorylation sites and is therefore constitutively active.8 Twenty-four hours later, the GFP+ DN3 thymocytes were sorted and analyzed for expression of indicated genes. After normalization to Hprt1, the expression of each gene in pMIG-infected cells was arbitrarily set to 1, and its relative expression in the presence of WT or mutant β-catenin was then calculated. Data were pooled from at least 3 independent experiments (n ≥ 7). Similar data were obtained with DN4 cells (not shown). (C) β-catenin–mediated Notch repression depends on TCF-1. DN3 thymocytes were sorted from control mice, Vav1-Cre Lef1−/−6 or Tcf7−/−, cultured in the presence of 5 µM MetBIO or BIO for 6 hours, and then harvested for gene expression analysis. After normalizing to Hprt1, the expression of each gene in MetBIO-treated cells was arbitrarily set to 1, and its relative expression in BIO-treated samples was then calculated. Data are pooled from 2 independent experiments (n ≥ 3). *, P < .05; **, P < .01; ***, P < .001 by Student t test. Note that although multiple TCF/LEF binding motifs were found within “–30 kb ∼ +10 kb” of transcription initiation sites of the Notch1, Dtx1, and Ptcra genes, we did not find enriched binding of TCF-1 to these motifs in DN3 thymocytes. Further studies are necessary to determine if repression of Notch1 and its targets by TCF-1 is mediated by direct regulation via more distal TCF/LEF motifs or by indirect mechanisms.
Our findings offer a unified answer to the 2 unresolved issues noted above. The answer being TCF-1 is responsible for early repression of Notch targets, including Ptcra, and hence attenuation of pre-TCR signaling, and is responsible for sustained repression of Notch1 after pre-TCR signals are diminished. Interestingly, TCF-1–mediated Notch1 downregulation is specific to thymocytes at the DN3 stage or beyond, where they are fully committed to the T-lineage, because Notch1 expression was not significantly affected by TCF-1 deficiency in DN1 cells and forced expression of the mutant β-catenin in DN1 did not repress Notch1 (supplemental Figure 1 on the Blood website). A requirement of β-catenin for normal thymopoiesis remains a contentious issue.9 It is therefore important to note that our gain-of-functional analysis demonstrates the sufficiency but not the necessity of activated β-catenin in repressing Notch signals in T-lineage–committed thymocytes. Nevertheless, our data revealed that the active Notch signaling is attenuated by a TCF-1-dependent mechanism during transition of DN1/early thymic progenitors to T-cell lineage-committed DN3 thymocytes. This model is consistent with the observations that Notch signaling is dispensable for late stages of thymocyte maturation1 and that ablation of both TCF-1 and LEF-1 arrests all thymocytes at the DN stage.6
The online version of this article contains a data supplement.
Authorship
Acknowledgments: The authors thank Hans Clevers for Tcf7−/− mice, Frank McCormick for the β-catenin constructs, and Juan C. Zuniga-Pflucker for the OP9-DL1 cells.
This study is supported by grants from the American Cancer Society (RSG-11-161-01-MPC [H.-H.X.]) and the National Institutes of Health (HL095540 [H.-H.X.]).
Contribution: S.Y. designed the study, performed research, and analyzed data; and H.-H.X. supervised research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Shuyang Yu, 2 Yuanmingyuan West Rd, Haidian District, Beijing 100193, People's Republic of China; e-mail: yushuyang@hotmail.com; and Hai-Hui Xue, 51 Newton Rd, BSB 3-772, Iowa City, IA 52242; e-mail: hai-hui-xue@uiowa.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal