Key Points
FLT3 activation cooperates with the MLL-AF4 fusion gene to fully abolish blood formation from hESCs.
FLT3 activation does not cooperate with the MLL-AF4 fusion oncogene to transform hESCs or hESC-derived hematopoietic progeny.
Mixed-lineage leukemia (MLL)–AF4 fusion arises prenatally in high-risk infant acute pro-B-lymphoblastic leukemia (pro-B-ALL). In human embryonic stem cells (hESCs), MLL-AF4 skewed hematoendothelial specification but was insufficient for transformation, suggesting that additional oncogenic insults seem required for MLL-AF4–mediated transformation. MLL-AF4+ pro-B-ALL expresses enormous levels of FLT3, occasionally because of activating mutations, thus representing a candidate cooperating event in MLL-AF4+ pro-B-ALL. Here, we explored the developmental impact of FLT3 activation alone, or together with MLL-AF4, in the hematopoietic fate of hESCs. FLT3 activation does not affect specification of hemogenic precursors but significantly enhances the formation of CD45+ blood cells, and CD45+CD34+ blood progenitors with clonogenic potential. However, overexpression of FLT3 mutations or wild-type FLT3 (FLT3-WT) completely abrogates hematopoietic differentiation from MLL-AF4–expressing hESCs, indicating that FLT3 activation cooperates with MLL-AF4 to inhibit human embryonic hematopoiesis. Cell cycle/apoptosis analyses suggest that FLT3 activation directly affects hESC specification rather than proliferation or survival of hESC-emerging hematopoietic derivatives. Transcriptional profiling of hESC-derived CD45+ cells supports the FLT3-mediated inhibition of hematopoiesis in MLL-AF4–expressing hESCs, which is associated with large transcriptional changes and downregulation of genes involved in hematopoietic system development and function. Importantly, FLT3 activation does not cooperate with MLL-AF4 to immortalize/transform hESC-derived hematopoietic cells, suggesting the need of alternative (epi)-genetic cooperating hits.
Introduction
Newborn cancer is progressively seen as a developmental biology disease.1 An intriguing newborn cancer is the infant pro-B/monocyte acute lymphoblastic leukemia (ALL) characterized by the hallmark genetic abnormality t(4;11) encoding the fusion mixed-lineage leukemia (MLL)-AF4, which is associated with a dismal prognosis and very brief latency, raising the question of how this disease evolves so quickly.2,-4 Compelling evidence indicates that MLL-AF4 arises prenatally during embryonic/fetal hematopoiesis.2,3,5,6 To understanding the developmental impact of MLL-AF4, we first need to elucidate which is the target cell for transformation and the mechanisms underlying MLL-AF4–mediated transformation.
MLL-AF4–induced leukemogenesis has been difficult to model, and bona fide MLL-AF4 disease models do not exist.7,,-10 Our understanding of MLL fusions comes from murine models, which do not recapitulate the human disease faithfully, suggesting that these mouse models may be missing essential components of leukemogenesis during early human development. It could be argued that the lack of an MLL-AF4 model may be because (1) a cell in a wrong developmental stage was targeted in the mouse; (2) the impact of other secondary hits has not been properly addressed; (3) MLL-AF4 requires the reciprocal AF4-MLL fusion protein to cause pro-B ALL as shown in the mouse; and (4) MLL-AF4 exerts its transforming function preferentially in human cells, indicating that the MLL-AF4 function has to be addressed using ontogenically primitive human stem cells. Among these, neonatal (cord blood [CB]–derived) CD34+ hematopoietic stem/progenitor cells (HSPCs) or prenatal (fetal- or embryonic-derived) cells represent ontogenically early candidate target cells. Thus, human embryonic stem cell (hESC)–derived hematopoietic differentiation constitutes a robust human-specific strategy to study the onset of hematopoiesis, representing a promising tool for modeling developmental mechanisms of human disease and lineage specification that cannot be addressed with patient samples or mouse models.11,12
We have reported that the expression of MLL-AF4 has a functional impact in CB−CD34+ HSPCs13 and hESC-hematopoietic cells.14 In CB−CD34+ HSPCs, MLL-AF4 enhanced hematopoietic engraftment and clonogenic potential but was not sufficient for leukemogenesis.13 In hESCs, MLL-AF4 altered the developmental cell fate, skewing the early hematoendothelial specification of hESCs.14 This mechanism suggests that the inability to develop an MLL-AF4 model is not the result of the human cell context or cell type targeted, but rather that additional oncogenic lesions are required for leukemogenesis.
Gene expression profiling showed that FLT3 is highly expressed in MLL-AF4+ pro-B-ALL. Moreover, it was shown that other MLL-rearranged leukemias display FLT3 mutations (FLT3-tyrosine kinase domain [TKD] or FLT3-internal tandem duplication [ITD]) in up to 20% of the cases, suggesting that they may represent candidate cooperating events.15,-17 Accordingly, Yamaguchi et al18 have reported that FLT3-TKD cooperates with MLL-AF4 to induce in vitro aggressive proliferation of the mouse cell line 32Dc. Several other groups suggest, however, that FLT3 mutations are not common in MLL-AF4+ pro-B-ALL, and that an increased transcriptional expression of FLT3 may act as a secondary cooperating hit.17,19,-21 The latter is supported by Guenther et al19 , who reported that FLT3 is a direct transcriptional target of MLL-AF4.
We have thus explored the developmental impact of FLT3 on its own, or in cooperation with MLL-AF4, in the hematopoietic fate of hESCs. We posed the following 3 questions. First, what is the developmental impact of FLT3 mutations/wild-type FLT3 (FLT3-WT) in the hematopoietic specification of hESCs? Second, do FLT3 mutations/FLT3-WT cooperate with MLL-AF4 during hematopoietic commitment of hESCs? Third, is expression of FLT3 mutations/FLT3-WT together with MLL-AF4 sufficient to confer a proliferative/survival advantage as anticipated for transforming oncogenes?
Materials and methods
Plasmid construction and lentiviral transduction
The MLL-AF4 complementary DNA (cDNA) (MLL-exon10 fused to AF4-exon8) and the FLT3-TKD (D835 mutation), FLT3-ITD, and FLT3-WT cDNAs were subcloned into the pRRL-EF1α-PGK-green fluorescent protein (GFP)/NEO vector.14 The following lentivectors were used: pRRL-EF1α-PGK-NEO (empty vector; EV), pRRL-EF1α-MLL-AF4-PGK-NEO (MLL-AF4), pRRL-EF1α-FLT3-TKD-PGK-GFP (FLT3-TKD), pRRL-EF1α-FLT3-ITD-PGK-GFP (FLT3-ITD), and pRRL-EF1α-FLT3-WT-PGK-GFP (FLT3-WT) (Figure 1A). Viral production and infection were performed as described previously.22 NEO-expressing hESCs were selected with G418 at 100 μg/mL for 3 weeks.22 In dual transfection experiments, FLT3/GFP-expressing viruses were used to infect G-418–resistant MLL-AF4–expressing hESCs. MLL-AF4 and FLT3 expression and mutational status were confirmed.
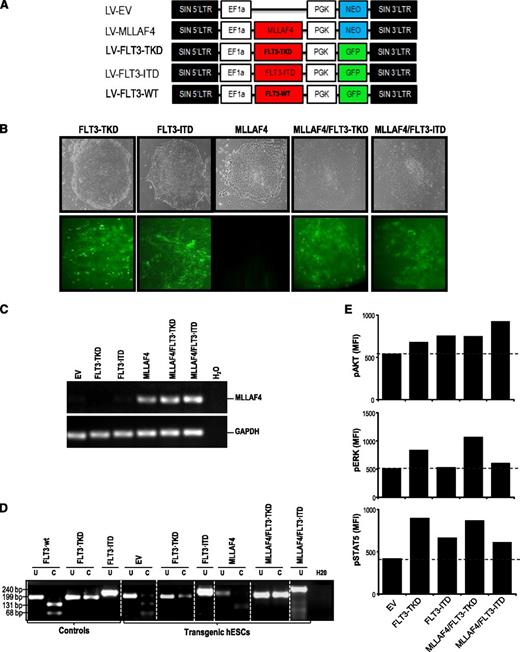
Generation of transgenic hESCs expressing MLL-AF4 and/or FLT3-activating mutations. (A) Schematic representation of the lentiviral vectors used. (B) Phase-contrast morphology (top) and fluorescence microscopy (bottom) of representative hESC colonies from FLT3-TKD-, FLT3-ITD-, MLL-AF4-, MLL-AF4/FLT3-TKD-, and MLL-AF4/FLT3-ITD-hESCs. (C) RT-PCR confirming expression of the MLL-AF4 transcript in transduced hESCs. GAPDH was used as a housekeeping gene. (D) PCR confirming the presence of either FLT3-TKD or FLT3-ITD mutations in transgenic hESCs. Vectors containing the FLT3 mutations were used as positive controls. (E) Phosphosignaling analysis of transduced hESCs, showing increased AKT, ERK, and STAT5 phosphorylation (relative to EV-hESCs) in cells transduced with FLT3-activating mutations.
Generation of transgenic hESCs expressing MLL-AF4 and/or FLT3-activating mutations. (A) Schematic representation of the lentiviral vectors used. (B) Phase-contrast morphology (top) and fluorescence microscopy (bottom) of representative hESC colonies from FLT3-TKD-, FLT3-ITD-, MLL-AF4-, MLL-AF4/FLT3-TKD-, and MLL-AF4/FLT3-ITD-hESCs. (C) RT-PCR confirming expression of the MLL-AF4 transcript in transduced hESCs. GAPDH was used as a housekeeping gene. (D) PCR confirming the presence of either FLT3-TKD or FLT3-ITD mutations in transgenic hESCs. Vectors containing the FLT3 mutations were used as positive controls. (E) Phosphosignaling analysis of transduced hESCs, showing increased AKT, ERK, and STAT5 phosphorylation (relative to EV-hESCs) in cells transduced with FLT3-activating mutations.
Human ESC culture
hESCs (AND-1 line) were maintained undifferentiated in a feeder-free culture as described.23,-25 Briefly, hESCs were cultured in Matrigel-coated T25 flasks in human feeders-conditioned medium supplemented with 4 ng/mL of basic fibroblast growth factor.26,-28 The medium was changed daily, and the cells were split weekly with 200U/mL of collagenase IV. This study was approved by The Comisión Nacional de Garantías del ISCIII y el Comité de Investigación de Preembriones Humanos la Consejería de Salud de la Junta de Andalucía to work with hESCs.
RNA extraction, cDNA synthesis, and MLL-AF4 and FLT3 gene expression
RNA extraction and reverse transcription were performed using the Europe Against Cancer group protocol.17 cDNA was used for conventional (MLL-AF4) and quantitative (FLT3) polymerase chain reaction (PCR). MLL-AF4 expression was confirmed using the following primers (Fw:5′-CAGGTCCAGAGCAGAGCAAAC-3′ and Rw:5′-GAGCACTTGGAGGTGCAGATG-3′) and PCR conditions (95°C for 10 minutes followed by 40 cycles of 95°C for 15 seconds and 60°C for 60 seconds).17 In quantitative PCR experiments, FLT3 expression was normalized to the expression of β-actin. Quantitative PCR was performed using SYBRGreen PCR Master mix (Applied BioSystem) and the 7500 reverse transcriptase (RT)-PCR system.
FLT3 gene mutation analysis
Flow cytometry characterization of hESCs
hESCs were dissociated with trypsin-EDTA, and the single-cell suspension was stained (2-5 × 105cells/mL) with TRA-1-60-PE, SSEA3-PE, SSEA4-fluorescein isothiocyanate (FITC), and OCT4-FITC (all from BD). Cells were then washed and stained with 7-amino-actinomycin D (7-AAD) for 15 minutes.25,30 Cells were analyzed using a FACS Canto-II-cytometer.
Expression of pluripotency-associated transcription factors
Transgenic hESCs were subjected to RT-PCR for detection of OCT4, NANOG, SOX2, and REX1 expression. GAPDH was used as a housekeeping gene. The primers and PCR conditions used were described elsewhere.31
In vivo teratoma formation
Animal protocols were approved by the local university hospital council on animal care and experimentation. Teratoma assay was conducted to confirm in vivo pluripotency.24,32 hESCs were implanted subcutaneously in 8-week-old immunodeficient mice (The Jackson Laboratory, Bar Harbor, ME). Teratoma growth was determined by palpation, and mice were euthanized 7 weeks after implantation. Teratomas were fixed, embedded in paraffin, and sections stained with hematoxylin/eosin. Immunocytochemistry analysis was performed for smooth muscle actin, pan-cytokeratin, α-fetoprotein, and β-III-tubulin (Dako).
Phosphoflow analysis of hESCs
For phosphosignaling, hESCs were incubated with TryplE (Invitrogen) for 5 minutes, washed and resuspended in staining buffer (BD Biosciences). Then, cells were fixed with Cytofix Buffer (BD Biosciences) and permeabilized in chilled Phosphoflow PermBuffer-III for 30 minutes before staining with anti-extracellular signal-regulated kinase (ERK)1/2 (pT302/pY204)-PE, anti-STAT5 (pY694)-PE, and anti-Akt (also known as protein kinase B; pS473). Data were analyzed in a FACSCanto-II-cytometer equipped with the FACSDiva software. The mean fluorescence intensity for each hESC line was compared with EV-hESC.
Immunoprecipitation and western blotting
Confluent hESCs were lysed in 1% Triton X-100 lysis buffer supplemented with Complete protease inhibitors cocktail and phosphatase inhibitors (Roche). Lysates were immunoprecipitated using protein-A agarose beads (Roche) and FLT3 antibody (clone S18; Santa Cruz). Immunoprecipitations were performed from 5-8 × 106 cells. Detection of tyrosine phosphorylation in FLT3 pull-downs was performed by immunoblotting using 4G10 antibody (Upstate). Signal was detected with the Odyssey infrared imaging system (Li-cor, Lincoln, NE). The MLL-rearranged THP1 line before and after FLT3L treatment was used as control.
Hematopoietic differentiation from hESCs
Undifferentiated hESCs were treated with collagenase IV and lifted of the Matrigel attachments. They were transferred to low-attachment plates to allow embryoid body (EB) formation in differentiation medium (KO-DMEM + 20% fetal bovine serum, 1% nonessential amino acids, 1mM of l-glutamine, 0.1 mM of β-mercaptoethanol). The medium was changed every 3 days and was supplemented with hematopoietic cytokines (300 ng/mL of SCF, 10 ng/mL of IL-3, 10 ng/mL of IL-6, and 50 ng/mL G-CSF, 25 ng/mL BMP-4). EBs were dissociated using collagenase B at days 4, 7, 11, and 15 of development. A single-cell suspension achieved by gentle pipetting and passage through a 70-μm cell strainer was stained with anti-CD31−PE, anti-CD45−APC, and anti-CD34−PECy7 (BD Biosciences) and 7-amino-actinomycin D (7AAD). Live cells (7AAD-) were analyzed using a FACSCanto-II-cytometer. Hemogenic precursors were identified as CD34+CD31+CD45−. Immature and total blood cells were identified as CD45+CD34+ and CD45+, respectively (Figure 2A).22,25
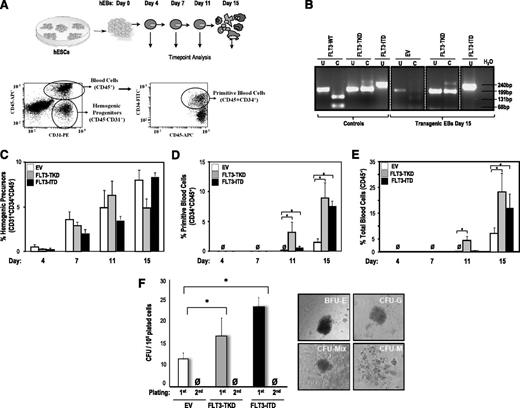
FLT3 activation enhances hematopoietic differentiation from hESCs. (A) Schematic of the hematopoietic differentiation of hESCs and end point analyses (top) and representative flow cytometry dot plots displaying how hemogenic precursors (CD45− CD31+), primitive blood cells (CD45+ CD34+), and total blood cells (CD45+) are identified (bottom). (B) PCR confirming the presence of either FLT3-TKD or FLT3-ITD mutations in transgenic day 15 hEBs. (C) Specification into hemogenic precursors is not significantly affected by FLT3 activation throughout EB development. However, FLT3 activation enhances hESC differentiation into primitive blood cells (D) and total blood cells (E). (F) CFU readout from d15 hEBs confirms a significant increase in hematopoietic potential in hESCs expressing either FLT3 mutation. Data are presented as mean ±SEM for 6 independent experiments. FLT3 mutation–expressing hESC-derived hematopoietic cells do not show stable in vitro replating efficiency in secondary CFU assays. Right panels depict representative CFU colonies.
FLT3 activation enhances hematopoietic differentiation from hESCs. (A) Schematic of the hematopoietic differentiation of hESCs and end point analyses (top) and representative flow cytometry dot plots displaying how hemogenic precursors (CD45− CD31+), primitive blood cells (CD45+ CD34+), and total blood cells (CD45+) are identified (bottom). (B) PCR confirming the presence of either FLT3-TKD or FLT3-ITD mutations in transgenic day 15 hEBs. (C) Specification into hemogenic precursors is not significantly affected by FLT3 activation throughout EB development. However, FLT3 activation enhances hESC differentiation into primitive blood cells (D) and total blood cells (E). (F) CFU readout from d15 hEBs confirms a significant increase in hematopoietic potential in hESCs expressing either FLT3 mutation. Data are presented as mean ±SEM for 6 independent experiments. FLT3 mutation–expressing hESC-derived hematopoietic cells do not show stable in vitro replating efficiency in secondary CFU assays. Right panels depict representative CFU colonies.
Colony-forming unit (CFU) assay
Gene expression profiling (GEP) and data analysis
CD45+ cells derived from EV-hESCs, MLL-AF4-hESCs, and MLL-AF4/FLT3-TKD-hESCs were sorted for GEP.14,33 RNA was isolated and its quality checked in the Agilent 2100 Bioanalyzer platform. RNA was labeled (Quick-Amp Labeling kit) with Cy3 and was hybridized with the Gene Expression Hybridization kit to a Whole Human Genome Microarray (G4112F; Agilent Technologies) following the manufacturer’s instructions.14,33 Four independent samples per condition were labeled and hybridized. Hierarchical clustering of genes and samples was performed with the 1− correlation metric and the unweighted average distance. A gene was considered differentially expressed when it was >2-fold up/down-regulated (P value < .01) compared with the control. Analysis of gene functions and canonic pathways significantly altered between experimental conditions was performed using the Ingenuity Pathway Analysis (IPA) (Ingenuity Systems, Redwood City, CA).14,34,35 Microarray data have been deposited in the National Center for Biotechnology Gene Expression Omnibus (NCBI-GEO) (GSE40103).
Cell cycle/apoptosis analysis of hemogenic precursors and CD45+ hematopoietic cells
EBs were dissociated at day 15 of development. Washed cells were incubated with anti-CD31−FITC, anti-CD34−FITC, anti-CD34−PECy7, and anti-CD45−APC. Cell cycle distribution and apoptosis status of hemogenic precursors and CD45+ blood cells were analyzed, as described previously in detail.14
Results
FLT3 activation alone or in combination with MLL-AF4 expression is compatible with hESC pluripotency and induces phosphorylation of FLT3R and FLT3-mediated signal transducing effectors.
To study the cooperation between MLL-AF4 and FLT3, FLT3-TKD and FLT3-ITD were subcloned in a lentivector expressing GFP (Figure 1A). Empty vector (EV)/NEO or MLL-AF4/NEO-hESCs had been established after G418 selection.14 G-418–resistant EV- and MLL-AF4–expressing hESCs were infected with viruses expressing FLT3-TKD/GFP or FLT3-ITD/GFP achieving >85% transduction efficiency (Figure 1B). Successful and stable expression of MLL-AF4 was confirmed by RT-PCR >20 passages after G418 selection (Figure 1C). FLT3 mutations were also confirmed by PCR (Figure 1D). Ectopic expression of FLT3 mutants or FLT3-WT gene induced FLT3 receptor phosphorylation (supplemental Figure 1A), and activated the 3 major intracellular pathways AKT, ERK, and STAT5.36 Phosphosignaling analysis showed increased phosphorylation of these signal-transducing effectors in hESCs transduced with FLT3 mutations (Figure 1E), confirming that FLT3 expression was functional.
Human ESC cultures were then analyzed for pluripotency markers and functional assays. hESCs transduced with FLT3 mutations retained expression of the antigens Tra-1-60, SSEA-3, SSEA-4, and OCT-4 (supplemental Figure 1B) and the pluripotency markers SOX2, NANOG, OCT4, and REX1 (supplemental Figure 1C). Functionally, all hESCs formed teratomas with 100% efficiency (supplemental Figure 1D), indicating that FLT3 activation is compatible with hESC pluripotency.
FLT3-activating mutations enhance hematopoietic differentiation of hESCs.
We tested whether FLT3-TKD and FLT3-ITD affect the hematopoietic cell fate of hESCs. During hEB differentiation, a population of primitive hemogenic precursors arises, which is responsible for hematopoietic and endothelial development.14,22,25,37 We investigated the effect of FLT3-TKD and FLT3-ITD on the emergence of hemogenic precursors (CD45−CD31+CD34+) throughout hEB development (days 4, 7, 11, and 15) (Figure 2A). We confirmed by PCR stable expression of ectopic FLT3 mutations on hEB differentiation (Figure 2B). Hemogenic precursors gradually emerge with time in all hESC lines studied, and their specification is not significantly affected by FLT3 mutations throughout development (Figure 2C).
We next assessed whether FLT3-TKD and FLT3-ITD influence subsequent hematopoietic commitment of hemogenic precursors. The emergence of primitive (CD45+CD34+) and total (CD45+) hematopoietic cells was analyzed throughout hEB development (Figure 2D-E). Blood cells did not emerge before day 10 of hEB development. Interestingly, both FLT3-TKD and FLT3-ITD enhanced (∼3- to 4-fold) hematopoietic differentiation of hESCs. FLT3-TKD accelerated the emergence of CD45+ and CD45+CD34+ blood cells (Figure 2D-E). Importantly, FLT3-TKD and FLT3-ITD also increased the clonogenic potential of hematopoietic progenitors derived from hEBs. Hematopoiesis generated from FLT3-activated hESCs displayed a 1.5- to 2-fold increased clonogenic potential (CFUs) in semisolid cultures (Figure 2F).
FLT3 cooperates with MLL-AF4 to abrogate hematopoietic commitment of hESCs.
The contribution of FLT3 to MLL-AF4–mediated hematopoiesis and leukemogenesis is controversial.17,18,38 We first explored whether FLT3-activating mutations cooperate with MLL-AF4 in the hematopoietic fate of hESCs. EV- and MLL-AF4-hESCs were transduced with either FLT3-TKD or FLT3-ITD. More than 85% of the cells within the EV/FLT3 mutation–expressing and MLL-AF4/FLT3 mutation–expressing hESC cultures were transduced as measured by GFP expression (Figure 1B); PCR for MLL-AF4 (Figure 1C) and FLT3-TKD or FLT3-ITD (Figure 1D); and phosphosignaling analysis for AKT, ERK, and STAT5 (Figure 1E).
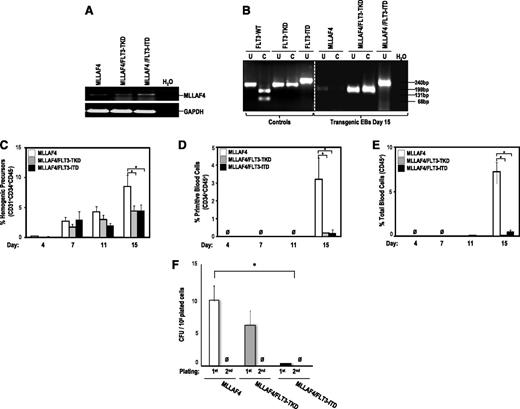
We investigated the effect of FLT3-TKD and FLT3-ITD in collaboration with MLL-AF4 on the emergence of hemogenic precursors throughout hEB development. We confirmed by PCR stable expression of ectopic FLT3 mutations and MLL-AF4 on hEB differentiation (Figure 3A-B). Both FLT3 mutations slightly impaired the specification into hemogenic precursors of MLL-AF4-hESCs (Figure 3C), whereas they completely blocked subsequent differentiation of MLL-AF4-hemogenic precursors into primitive (Figure 3D) and total blood cells (Figure 3E). FLT3-TKD and FLT3-ITD also abrogated the clonogenic potential of MLL-AF4–expressing hematopoietic derivatives (Figure 3F). These data indicate that MLL-AF4 inhibits the FLT3-TKD/ITD–mediated enhanced blood differentiation of hESCs.
FLT3-activating mutations block hematopoietic differentiation from MLL-AF4-hESCs. (A) RT-PCR confirming stable expression of MLL-AF4 in transgenic day 15 hEBs. (B) PCR confirming the presence of either FLT3-TKD or FLT3-ITD mutations in transgenic day 15 hEBs. FLT3-activating mutations only impair late specification into hemogenic precursors of MLL-AF4–expressing hESCs (C) but completely block differentiation of MLL-AF4–expressing hESCs into primitive blood cells (D) and total blood cells (E). (F) CFU readout from d15 hEBs confirms a significant decrease in hematopoietic potential in hESCs coexpressing MLL-AF4 and FLT3 mutations. Data are presented as mean ± SEM for 9 independent experiments. MLL-AF4/FLT3 mutation–expressing hESC-derived hematopoietic cells do not show stable in vitro replating efficiency in secondary CFU assays.
FLT3-activating mutations block hematopoietic differentiation from MLL-AF4-hESCs. (A) RT-PCR confirming stable expression of MLL-AF4 in transgenic day 15 hEBs. (B) PCR confirming the presence of either FLT3-TKD or FLT3-ITD mutations in transgenic day 15 hEBs. FLT3-activating mutations only impair late specification into hemogenic precursors of MLL-AF4–expressing hESCs (C) but completely block differentiation of MLL-AF4–expressing hESCs into primitive blood cells (D) and total blood cells (E). (F) CFU readout from d15 hEBs confirms a significant decrease in hematopoietic potential in hESCs coexpressing MLL-AF4 and FLT3 mutations. Data are presented as mean ± SEM for 9 independent experiments. MLL-AF4/FLT3 mutation–expressing hESC-derived hematopoietic cells do not show stable in vitro replating efficiency in secondary CFU assays.
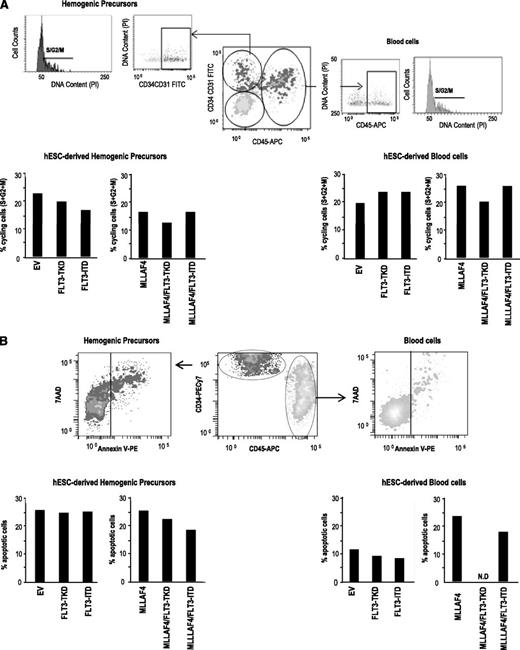
The developmental impact of FLT3-TKD and FLT3-ITD in hematopoietic cell fate of hESCs may be the result of (1) direct regulation of hematopoietic specification of hESCs or (2) effects on the proliferation or survival of the emerging hematopoietic cells. To address this, cell cycle distribution (Figure 4A) and apoptosis (Figure 4B) were analyzed within the hESC-derived hemogenic precursor population and within the CD45+ blood cells (Figure 4A-B). No differences in the proportion of cycling (Figure 4A) or apoptotic (Figure 4B) hESC-derived hemogenic precursors or blood cells were observed when FLT3-TKD or FLT3-ITD was expressed alone or together with MLL-AF4. This suggests that FLT3-activating mutations promote hESC hematopoietic specification rather than selective proliferation or survival of hESC-emerging hematopoietic derivatives.
Cell cycle and apoptosis analysis reveal that the effect of FLT3 mutations alone or in combination with MLL-AF4 on hESC hematopoietic specification is independent of proliferation or survival. (A) Top, representative flow cytometric analysis showing how cell cycle analysis was analyzed in hemogenic precursors and hematopoietic cells. Bottom, similar proportion of cycling (S+G2+M) hemogenic precursors (left) and hematopoietic cells (right) among the distinct transgenic hESCs. (B) Top, representative flow cytometric analysis showing how apoptosis was analyzed in hemogenic precursors and hematopoietic cells. Bottom, similar proportion of apoptotic (Annexin V+) hemogenic precursors (left) and hematopoietic cells (right) among the distinct transgenic hESCs. N.D: not determined because of the complete absence of CD45+ cells for analysis.
Cell cycle and apoptosis analysis reveal that the effect of FLT3 mutations alone or in combination with MLL-AF4 on hESC hematopoietic specification is independent of proliferation or survival. (A) Top, representative flow cytometric analysis showing how cell cycle analysis was analyzed in hemogenic precursors and hematopoietic cells. Bottom, similar proportion of cycling (S+G2+M) hemogenic precursors (left) and hematopoietic cells (right) among the distinct transgenic hESCs. (B) Top, representative flow cytometric analysis showing how apoptosis was analyzed in hemogenic precursors and hematopoietic cells. Bottom, similar proportion of apoptotic (Annexin V+) hemogenic precursors (left) and hematopoietic cells (right) among the distinct transgenic hESCs. N.D: not determined because of the complete absence of CD45+ cells for analysis.
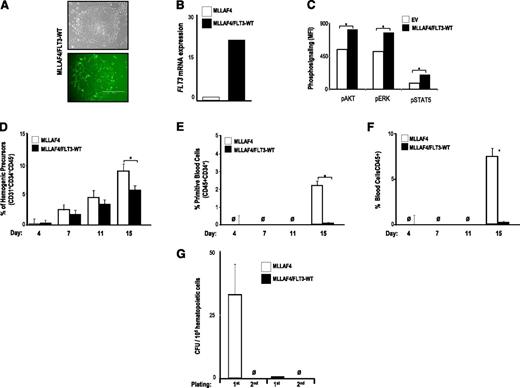
Several groups have reported the presence of FLT3-TKD mutations in MLL-rearranged ALLs,15,16,39 whereas others have shown that FLT3 mutations in MLL-rearranged ALLs do not occur.17,20,21,40,41 High expression of FLT3 is a hallmark step in the pathogenesis of MLL-AF4+ ALL,17 but it has not been resolved whether the constitutive activation of FLT3 in MLL-AF4+ ALL is because of the presence of activating FLT3 mutations or simply the result of an increased expression of FLT3. We thus investigated whether ectopic expression of FLT3-WT cooperates with MLL-AF4 in the hematopoietic fate of hESCs. MLL-AF4-hESCs were transduced with FLT3-WT, and >90% of the cells within the MLL-AF4/FLT3-WT-hESC cultures were transduced as measured by GFP expression (Figure 5A); PCR for FLT3 (Figure 5B); and phosphosignaling analysis for AKT, ERK, and STAT5 (Figure 5C). Ectopic expression of FLT3-WT barely impaired specification into hemogenic precursors of MLL-AF4-hESCs (Figure 5D), but it completely blocked differentiation of MLL-AF4-hemogenic precursors into primitive (Figure 5E) and total blood cells (Figure 5F). Additionally, FLT3-WT overexpression fully abrogated the clonogenic potential of MLL-AF4–expressing hESC-hematopoietic derivatives (Figure 5G). This finding indicates that FLT3 activation, either through activating mutations or overexpression of the WT gene, abolishes hematopoietic differentiation of MLL-AF4-hESCs.
FLT3 WT overexpression also abolishes hematopoietic differentiation from MLL-AF4-hESCs. (A) Phase-contrast morphology (top) and fluorescence microscopy (bottom) of colonies from MLL-AF4/FLT3-WT-hESCs. (B) Quantitative RT-PCR confirming efficient transduction and stable expression of FLT3-WT. (C) Phosphosignaling analysis of transduced hESCs, showing increased AKT, ERK, and STAT5 phosphorylation in cells transduced with FLT3-WT. FLT3-WT slightly impairs specification into hemogenic precursors of MLL-AF4–expressing hESCs (D), whereas it completely abolishes differentiation of MLL-AF4–expressing hESCs into primitive blood cells (E) and total blood cells (F). (G) CFU readout from day 15 hEBs confirms a significant decrease in hematopoietic potential in hESCs coexpressing MLL-AF4 and FLT3-WT. Data are presented as mean ± SEM for 5 independent experiments.
FLT3 WT overexpression also abolishes hematopoietic differentiation from MLL-AF4-hESCs. (A) Phase-contrast morphology (top) and fluorescence microscopy (bottom) of colonies from MLL-AF4/FLT3-WT-hESCs. (B) Quantitative RT-PCR confirming efficient transduction and stable expression of FLT3-WT. (C) Phosphosignaling analysis of transduced hESCs, showing increased AKT, ERK, and STAT5 phosphorylation in cells transduced with FLT3-WT. FLT3-WT slightly impairs specification into hemogenic precursors of MLL-AF4–expressing hESCs (D), whereas it completely abolishes differentiation of MLL-AF4–expressing hESCs into primitive blood cells (E) and total blood cells (F). (G) CFU readout from day 15 hEBs confirms a significant decrease in hematopoietic potential in hESCs coexpressing MLL-AF4 and FLT3-WT. Data are presented as mean ± SEM for 5 independent experiments.
FLT3 activation does not cooperate with MLL-AF4 for in vitro transformation of hESC-derived hematopoietic cells.
We have reported previously that MLL-AF4 on its own is not sufficient for in vitro transformation of hESC-derived hematopoietic cells14 and that secondary oncogenic hits may be required, making FLT3 activation a candidate.15,,-18,21,39,42 The data presented here reveal that expression of the MLL-AF4 fusion oncogene together with FLT3 mutations does not confer either an in vitro proliferative or survival advantage to hESC-derived hemogenic precursors nor to CD45+ hematopoietic cells (Figure 4A-B). Functionally, FLT3 activation alone (Figure 2F) or in combination with MLL-AF4 expression (Figures 3F and 5G) does not confer stable in vitro replating efficiency of hematopoietic cells in CFU assays, confirming that FLT3 mutations are not secondary or cooperating hits for MLL-AF4 in the transformation of embryonic hematopoietic cells.
Transcriptional changes underlie the inhibition of hematopoiesis mediated by FLT3-TKD in MLL-AF4–expressing hESC-derived CD45+ hematopoietic cells.
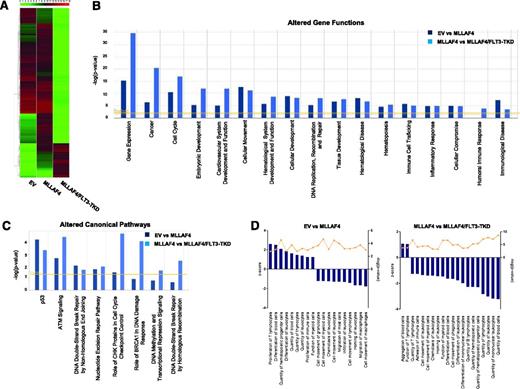
To identify patterns of gene expression that could explain molecularly the developmental impact of FLT3-TKD in MLL-AF4–expressing hESC-derived CD45+ hematopoietic cells, we next performed a microarray analysis. A heatmap representation of global gene expression changes indicates that FLT3-TKD acts as a global transcriptional repressor when overexpressed in MLL-AF4–expressing hESC-derived CD45+ cells (Figure 6A). The genes differentially expressed in MLL-AF4 vs EV hESC-derived CD45+ cells and MLL-AF4/FLT3-TKD vs MLL-AF4 hESC-derived CD45+ cells were analyzed at the functional level using the IPA software. Analysis of the altered genes revealed that several gene functions (Figure 6B) and canonic pathways (Figure 6C) were altered in both MLL-AF4 vs EV and MLL-AF4/FLT3-TKD vs MLL-AF4 hESC-derived CD45+ cells. Among the altered gene functions, gene expression, cancer, embryonic development, hematologic system development and function, and hematopoiesis are the most significantly modulated cell functions on the expression of MLL-AF4 or MLL-AF4 together with FLT3-TKD (Figure 6B). Those genes differentially regulated in MLL-AF4 (vs EV) hESC-derived CD45+ cells, and in MLL-AF4/FLT3-TKD (vs MLL-AF4) hESC-derived CD45+ cells, which were classified by the IPA software as involved in hematopoietic system development and function, were analyzed in more detail (Figure 6D). MLL-AF4 expression enhances several hematopoietic functions (positive z score) mainly related to proliferation and differentiation of hematopoietic cells, while inhibiting (negative z score) biofunctions linked to cell movement, migration, and chemotaxis. Interestingly, ectopic expression of FLT3-TKD in MLL-AF4–expressing hESC-derived CD45+ cells has a strong impact on these biofunctions associated with a marked inhibition (negative z score) of biofunctions involved in the physiology and homeostasis of blood cell differentiation, and function (Figure 6D). Overall, this analysis indicates that transcriptional changes underlie the inhibition of hematopoiesis on FLT3-TKD expression in MLL-AF4–expressing hESC-derived CD45+ hematopoietic cells.
Gene expression profiling identifies gene functions and signaling pathways altered in MLL-AF4 and MLL-AF4/FLT3-TKD hESCs-derived CD45+ hematopoietic cells. (A) Heatmap diagram depicting the global gene expression profiling for EV-, MLL-AF4-, and MLL-AF4/FLT3-TKD hESC-CD45+ blood cells. The upper color bar codifies the gene expression in a log2 scale. Expression levels vary from highly expressed (red) to nonexpressed (green) genes. (B-D) After gene expression microarray analysis, the groups of genes differentially expressed (P value < .01; 2-fold regulation) in MLL-AF4 vs EV and MLL-AF4/FLT3-TKD vs MLL-AF4 hESC-CD45+ cells were compared, and the lists of gene functions and canonic pathways significantly altered were generated using the IPA software. IPA software–based data mining generated a list of significantly modulated gene functions (B) and canonic pathways (C) in MLL-AF4 vs EV hESC-CD45+ cells and MLL-AF4/FLT3-TKD vs MLL-AF4 hESC-CD45+ cells. (D) A more profound analysis was then performed for all of the genes classified by the IPA software as involved in hematopoietic system development and function/hematopoiesis. The IPA analysis–based z score is an estimation of the activation/inhibition status of a given category within an altered gene function.
Gene expression profiling identifies gene functions and signaling pathways altered in MLL-AF4 and MLL-AF4/FLT3-TKD hESCs-derived CD45+ hematopoietic cells. (A) Heatmap diagram depicting the global gene expression profiling for EV-, MLL-AF4-, and MLL-AF4/FLT3-TKD hESC-CD45+ blood cells. The upper color bar codifies the gene expression in a log2 scale. Expression levels vary from highly expressed (red) to nonexpressed (green) genes. (B-D) After gene expression microarray analysis, the groups of genes differentially expressed (P value < .01; 2-fold regulation) in MLL-AF4 vs EV and MLL-AF4/FLT3-TKD vs MLL-AF4 hESC-CD45+ cells were compared, and the lists of gene functions and canonic pathways significantly altered were generated using the IPA software. IPA software–based data mining generated a list of significantly modulated gene functions (B) and canonic pathways (C) in MLL-AF4 vs EV hESC-CD45+ cells and MLL-AF4/FLT3-TKD vs MLL-AF4 hESC-CD45+ cells. (D) A more profound analysis was then performed for all of the genes classified by the IPA software as involved in hematopoietic system development and function/hematopoiesis. The IPA analysis–based z score is an estimation of the activation/inhibition status of a given category within an altered gene function.
These transcriptional changes support that FLT3-TKD cooperates with MLL-AF4 to abrogate the hematopoietic commitment of hESCs. However, FLT3-TKD did not cooperate with MLL-AF4 to confer in vitro or in vivo transformation of hESCs/hESC-derived hematopoietic cells. Guenther et al19 recently reported 159 MLL-AF4 direct target genes identified in leukemic samples, many of which are key for hematopoietic stem cell identity and self-renewal in human leukemia. We thus analyzed our gene expression data to determine whether these specific MLL-AF4 target genes are modulated in our cellular system. Ectopic expression of MLL-AF4 alone or together with FLT3-TKD induces transcriptional changes in subsets of this gene list in hESC-derived CD45+ cells (Figure 7A). Specifically, 75 and 106 MLL-AF4 target genes are transcriptionally modulated on expression of MLL-AF4 alone or MLL-AF4 along with FLT3-TKD, respectively (Figure 7B). These MLL-AF4 target genes differentially expressed in MLL-AF4 vs EV and MLL-AF4/FLT3-TKD vs MLL-AF4 hESC-derived CD45+ cells were analyzed using IPA. We found that MLL-AF4 target genes regulated by MLL-AF4 alone did not enhance biofunctions related to transformation. However, the subset of MLL-AF4 target genes regulated by expression of FLT3-TKD in MLL-AF4–expressing hESC-derived CD45+ hematopoietic cells is associated with an enhancement in biofunctions related to leukemia and cell transformation (positive z score) (Figure 7C-D).
Comparative analysis of the MLL-AF4 target genes reported by Guenther et al19 in MLL-AF4 and MLL-AF4/FLT3-TKD hESC-derived CD45+ hematopoietic cells. (A) Heatmap diagram depicting the expression of the MLL-AF4 target genes reported by Guenther et al19 for EV-, MLL-AF4-, and MLL-AF4/FLT3-TKD hESC-CD45+ blood cells. The upper color bar codifies the gene expression in a log2 scale. Expression levels vary from highly expressed (red) to nonexpressed (green) genes. (B) Number of MLL-AF4 target genes (of the159 reported by Guenther et al19 ) found differentially expressed in MLL-AF4 vs EV hESC-CD45+ cells and MLL-AF4/FLT3-TKD vs MLL-AF4 hESC-CD45+. (C) The MLL-AF4 target genes reported by Guenther et al19 differentially expressed in MLL-AF4 vs EV hESC-CD45+ cells (left panel) and MLL-AF4/FLT3-TKD vs MLL-AF4 hESC-CD45+ (right panel) were classified by IPA software. The z score is an estimation of the activation/inhibition status of a given category within an altered gene function. (D) List of MLL-AF4 target genes differentially expressed between MLL-AF4/FLT3-TKD and MLL-AF4 hESC-CD45+ cells.
Comparative analysis of the MLL-AF4 target genes reported by Guenther et al19 in MLL-AF4 and MLL-AF4/FLT3-TKD hESC-derived CD45+ hematopoietic cells. (A) Heatmap diagram depicting the expression of the MLL-AF4 target genes reported by Guenther et al19 for EV-, MLL-AF4-, and MLL-AF4/FLT3-TKD hESC-CD45+ blood cells. The upper color bar codifies the gene expression in a log2 scale. Expression levels vary from highly expressed (red) to nonexpressed (green) genes. (B) Number of MLL-AF4 target genes (of the159 reported by Guenther et al19 ) found differentially expressed in MLL-AF4 vs EV hESC-CD45+ cells and MLL-AF4/FLT3-TKD vs MLL-AF4 hESC-CD45+. (C) The MLL-AF4 target genes reported by Guenther et al19 differentially expressed in MLL-AF4 vs EV hESC-CD45+ cells (left panel) and MLL-AF4/FLT3-TKD vs MLL-AF4 hESC-CD45+ (right panel) were classified by IPA software. The z score is an estimation of the activation/inhibition status of a given category within an altered gene function. (D) List of MLL-AF4 target genes differentially expressed between MLL-AF4/FLT3-TKD and MLL-AF4 hESC-CD45+ cells.
It was reported that inhibition of DNA damage response (DDR) barriers accelerated MLL-rearrangement leukemogenesis.43 Our GEP reveals that several pathways involved in DDR are affected by expression of MLL-AF4 alone or in conjunction with FLT3-TKD (Figure 6C). We found a global downregulation of many components associated with DDR signaling pathways such as DNA repair by homologous recombination, nonhomologous end-joining, or ATM signaling (supplemental Figure 2A-C). This finding supports the observed FLT3-mediated inhibition of hematopoiesis in MLL-AF4–expressing hESCs and indicates that although FLT3-TKD does not cooperate with MLL-AF4 to confer functional transformation of hESCs or hESC-derived cells, it induces transcriptional modulation of MLL-AF4 target genes in MLL-AF4/FLT3-TKD hESCs-derived CD45+ hematopoietic cells.
Discussion
MLL-AF4+ ALL is a dismal leukemia that manifests in the first year of life.6 MLL-AF4 is the initiating leukemogenic event with an in utero origin.6 Our understanding about how MLL-AF4 deregulates early hematopoietic development is very limited, because despite recent advances in studies on mice7,9 and human cellular systems,13,14 current models do not recapitulate the disease phenotype or latency. Studies using cells from patients with MLL-AF4 are unable to address the developmental genesis of the hematopoietic system. Biphenotypic MLL-AF4+ infant ALL harbors and expresses fusion both in B cells and monocytes, suggesting that MLL-AF4 may arise in an ontogenically early hematopoietic stem cell.3 We recently reported that MLL-AF4 affects the developmental fate of human CB−CD34+ hematopoietic stem cells and hESCs but was insufficient for leukemogenesis, suggesting that additional cooperating oncogenic hits are required for MLL-AF4–mediated leukemogenesis.13,14
Despite current controversy about the presence of FLT3 gene mutations in MLL-AF4+ ALL,15,-17,20,21,39 GEP identified the FLT3 gene consistently highly expressed in MLL-AF4+ ALL, indicating that FLT3 activation may represent a candidate-cooperating lesion in MLL-AF4–mediated ALL. hESCs and hESC derivatives enable the study of early human development that cannot otherwise be addressed by patient sample analyses or mouse models. Hematopoietic differentiation of hESCs represents a strategy to study the onset of hematopoiesis, particularly the emergence of the earliest events leading to specification of the hematopoiesis.11,12 We have thus explored the role of FLT3 alone, or in cooperation with MLL-AF4, in the hematopoietic fate of hESCs and hESC-hemogenic precursors.
Activation of FLT3 in hESCs promoted hematopoietic specification. Opposite to CD45+ blood commitment, FLT3 mutations exerted neither blockage nor promotion of hESC specification into hemogenic precursors. These data suggest a dispensable role of FLT3 in the specification of human hemogenic endothelium. In fact, GEP revealed that FLT3 is expressed in hESC-derived CD45+ blood cells but not in CD45−CD31+ hemogenic precursors.
FLT3-activating mutations affect myeloid vs lymphoid lineage restriction of adult HSPCs.44 FLT3 also plays a key role in controlling the homeostasis of B-cell development; it is highly expressed in hematopoietic stem cells and pro-B cells but becomes down-regulated at the pre-B-cell stage. This is in keeping with the accumulation of FLT3 expression in patients with MLL-AF4+ who display a differentiation block at the pro-B-cell stage. In human embryonic hematopoiesis, cell cycle or apoptosis did not account for the enhanced hematopoiesis found on FLT3 activation, indicating that FLT3-mediated effects underlie hESC specification rather than selective proliferation or survival of hESC-emerging hematopoietic derivatives. This mechanism suggests that, signaling downstream, the activated receptor may differ between embryonic and adult/definitive hematopoiesis. Although FLT3-TKD and FLT3-ITD mutations have been reported to signal through distinct mechanisms, their ectopic expression in hESCs induced similar phosphorylation of the signal-transducing effectors AKT, ERK, and STAT5. Activation of FLT3 in hESCs was compatible with pluripotency, in line with previous findings supporting the importance of AKT and ERK in hESC maintenance.45
Intriguingly, FLT3 activation in MLL-AF4-hESCs robustly abrogated hematopoietic commitment of hESCs but barely affected MLL-AF4-hESC specification into hemogenic precursors, indicating that MLL-AF4 cooperates with FLT3 to abolish hematopoietic commitment of hESCs. Similarly, Furuichi et al38 have reported that FLT3 activation antagonizes with MLL rearrangements to inhibit proliferation of MLL-rearranged cell lines. Additionally, Sexauer et al46 have recently shown that FLT3-ITD blocks terminal myeloid differentiation. In MLL-AF4-hESCs, activation of FLT3 blocked hematopoietic commitment without affecting proliferation or survival of hESC-emerging hematopoietic derivatives. GEP analysis revealed that expression of FLT3-TKD in MLL-AF4–expressing hESC-derived CD45+ cells resulted in a genetic profile consistent with the inhibition of biofunctions involved in the physiology and homeostasis of blood cell differentiation and function, indicating that transcriptional changes underlie the inhibition of hematopoiesis on FLT3-TKD expression in MLL-AF4–expressing hESC-derived CD45+ hematopoietic cells. Similar to what occurs in primary MLL-AF4+ ALL, MLL-AF4–expressing hESC-derived CD45+ cells expressed high levels of MYC, HOXA9, SET, and RAN, which are proto-oncogenes activated by MLL-AF4 via aberrant H3K79 dimethylation and DOT1L recruitment.47
Coexpression of both MLL-AF4 and FLT3 mutations did not immortalize hESCs or hESC-derived hematopoietic derivatives. hESCs and hESC hematopoietic cells coexpressing both MLL-AF4 and FLT3 mutations did not display a proliferative and/or survival advantage and failed to confer replating efficiency in hematopoietic CFU assays. This finding suggests that either additional oncogenic hits7 may be required for leukemogenesis, or that embryonic cells are not the appropriate cellular target for MLL-AF4–mediated transformation. It cannot be ruled out that other embryonic precursors or fetal HSPCs represent better candidate target cells in which MLL-AF4 originates and/or exerts its oncogenic function. Yamaguchi et al18 reported that MLL-AF4 together with FLT3-TKD conferred the mouse cell line 32Dc IL3-independent robust proliferative capacity, supporting the need for further (epi)-genetic oncogenic events.
Guenther et al19 reported 159 MLL-AF4 direct target genes. Analysis of these 159 MLL-AF4 target genes in our GEP datasheet shows that 75 and 106 MLL-AF4 target genes are transcriptionally modulated on expression of MLL-AF4 alone or MLL-AF4 along with FLT3-TKD, respectively. These genes differentially expressed in MLL-AF4/FLT3-TKD hESC-derived blood cells were analyzed using IPA software, and it was found that expression of FLT3-TKD in MLL-AF4–expressing hESC-derived CD45+ cells regulates a subset of MLL-AF4 target genes involved in cell transformation and leukemia. Additionally, it has been reported that DDR is a rate-limiting event for acquisition of stem cell properties in MLL-mediated transformation, as experimental inhibition of the DDR barrier accelerated leukemia development.43 Our GEP reveals a global downregulation of many components associated with DDR master signaling pathways. These GEP data indicate that although FLT3-TKD does not cooperate with MLL-AF4 to transform hESCs or hESC-derived hematopoietic cells, it induces transcriptional modulation of MLL-AF4 target genes in hESCs-derived CD45+ hematopoietic cells, partially resembling the scenario found by Guenther et al19 by ChIP-Seq analysis.
Epigenetics represent a key player in MLL-AF4–driven transformation.48,49 In contrast to many human cancers, which are commonly characterized by hypomethylation in nonpromoter regions, MLL-AF4+ ALL displays genome-wide hypermethylation at nonpromoter sequences.48,49 Global hypomethylation is usually associated with genomic instability, allowing additionally acquired genetic hits to propel a premalignant clone into a fully transformed state. The global hypermethylation of MLL-AF4+ ALL might explain why additional genetic lesions have not been discovered in MLL-rearranged ALL.40,41
We provide a hESC model to deepen our understanding about the mechanisms underlying the developmental impact of the MLL-AF4 and FLT3 signaling pathway during human early embryonic development. It also offers a unique in vitro system in which to test the ability of potential cooperating oncogenic events (reciprocal AF4-MLL that causes epigenetic remodeling and strong transcriptional enhancement)50 or causal genotoxic compounds to induce leukemic transformation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was funded by FIS/FEDER (PI10/00449 to P.M. and PI11/00119 to C.B.) and by The Spanish Association Against Cancer to P.M. D.R.-M. is supported by PFIS scholarship (FI11/00511). C.B., P.J.R., and V.R.-M. are supported by “Miguel Servet” Fellowships (CP07/0059, CP09/0063, and CP12/03175). R.M. is supported by the ISCIII (CA10/01332). P.M. is an ICREA investigator supported by the ISCIII Red de Terapia Celular (Tercel, RD12/0019/0006).
Authorship
Contribution: C.B. and P.M. designed the research of this manuscript. C.B., V.A., R.M., O.N.-M., D.R.-M., V.R.-M., P.J.R., and M.J.A.-B. performed research and analyzed data. C.B. and P.M. wrote the manuscript. The manuscript has been seen and approved by all authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Clara Bueno, Centre for Genomics and Oncological Research: Pfizer/University of Granada/Andalusian Government, Avda de la Ilustración 114, 18016, Granada, Spain; e-mail: clara.bueno@genyo.es; and Pablo Menendez, Centre for Genomics and Oncological Research: Pfizer/University of Granada/Andalusian Government, Avda de la Ilustración 114, 18016, Granada, Spain, and Josep Carreras Leukemia Research Institute-Facultat de Medicina, University of Barcelona, Instituciò Catalana de Reserca i Estudis Avançats Carrer Casanova 143, Barcelona 08036, Spain. e-mail: pablo.menendez@genyo.es.