Key Points
NRP1 promotes brain angiogenesis cell autonomously in endothelium, independently of heterotypic interactions with nonendothelial cells.
NRP1 plays a key role in endothelial tip rather than stalk cells during vessel sprouting in the brain.
Abstract
Neuropilin (NRP) 1 is a receptor for the vascular endothelial growth factor (VEGF)-A and is essential for normal angiogenesis. Previous in vitro experiments identified NRP1 interactions with VEGF-A’s main signaling receptor VEGFR2 within endothelial cells, but also between nonendothelial NRP1 and endothelial VEGFR2. Consistent with an endothelial role for NRP1 in angiogenesis, we found that VEGFR2 and NRP1 were coexpressed in endothelial tip and stalk cells in the developing brain. In addition, NRP1 was expressed on two cell types that interact with growing brain vessels—the neural progenitors that secrete VEGF-A to stimulate tip cell activity and the pro-angiogenic macrophages that promote tip cell anastomosis. Selective targeting of Nrp1 in each of these cell types demonstrated that neural progenitor- and macrophage-derived NRP1 were dispensable, whereas endothelial NRP1 was essential for normal brain vessel growth. NRP1 therefore promotes brain angiogenesis cell autonomously in endothelium, independently of heterotypic interactions with nonendothelial cells. Genetic mosaic analyses demonstrated a key role for NRP1 in endothelial tip rather than stalk cells during vessel sprouting. Thus, NRP1-expressing endothelial cells attained the tip cell position when competing with NRP1-negative endothelial cells in chimeric vessel sprouts. Taken together, these findings demonstrate that NRP1 promotes endothelial tip cell function during angiogenesis.
Introduction
In vertebrates, organ development, homeostasis, and repair rely on properly perfused blood vessel networks. The first blood vessels in the embryo are assembled from single-cell precursors, which coalesce into a honeycomb-shaped vascular plexus in a process known as vasculogenesis.1 This primitive plexus then expands through sprouting and remodeling to supply all tissues and organs in a mechanism termed angiogenesis.1 Angiogenesis is normally quiescent in adults but can be reactivated, for example during tissue repair in wound healing, in ischemic eye disease, or during solid tumor growth.2
Angiogenesis depends on the vascular endothelial growth factor (VEGF)-A, which binds several different receptors on endothelial cells.1,3 The tyrosine kinase receptor VEGFR2, also known as FLK1 in mouse and KDR in humans, is the main signal transducing VEGF-A receptor in endothelial cells; it stimulates endothelial cell proliferation and migration through a plethora of downstream signaling events, and loss of VEGFR2 therefore halts blood vessel formation by vasculogenesis early during embryogenesis.4 After vasculogenesis, VEGFR2 promotes angiogenesis by acting in the specialized endothelial tip cells that head vascular sprouts and extend filopodia to sense growth factor gradients composed of VEGF-A isoforms with different matrix affinities.5,6 Consistent with an important role for VEGFR2 in endothelial guidance, high levels of VEGFR2 confer a competitive advantage to endothelial cells as they negotiate the tip cell position with their neighboring cells.7
In cultured human endothelial cells, VEGFR2 forms complexes with NRP1, a nontyrosine kinase receptor for the VEGF165 isoform of VEGF-A.8,9 Complex formation is thought to promote VEGF165 signaling through VEGFR2 and activate signaling pathways involved in cell migration and angiogenic sprout formation.8-11 The VEGF165-mediated interaction between NRP1 and VEGFR2 may occur when both receptors are coexpressed on the same endothelial cell or on neighboring endothelial cells (homotypic interaction).9 Alternatively, NRP1 may be expressed on nonendothelial cells that interact with VEGFR2-expressing endothelial cells (heterotypic interaction). For example, it has been proposed that endothelial VEGFR2 interacts with tumor cell NRP1 in trans to stimulate VEGFR2 phosphorylation.9 Consistent with the idea of heterotypic roles for NRP1 in angiogenesis, treatment with a soluble dimerized form of NRP1 can rescue defective vascular outgrowth from paraaortic splanchnopleural explants of NRP1-deficient mouse embryos in vitro.12 However, the relevance of nonendothelial NRP1 for angiogenesis in vivo was not established in these studies.
In the mouse, NRP1 overexpression leads to the formation of excess blood vessels that are leaky and hemorrhagic, causing lethality by embryonic day (E) 17.5.13 Vice versa, the targeted disruption of Nrp1 in mice reduces blood vessel growth and causes embryonic death even earlier, between E10.5 and E14.5, depending on the genetic background.14-16 Vascular defects are particularly severe in the brain and spinal cord.14,17 Accordingly, the developing nervous system provides a good model to study NRP1 function in angiogenesis. Strikingly, NRP1 is expressed not only by endothelial cells in the brain, but also by the neural progenitors that secrete VEGF-A to promote vessel sprouting into the brain18-20 and regulate angiogenesis via expression of beta-8 integrin.21 Moreover, NRP1 is expressed by proangiogenic tissue macrophages that are the precursors of brain microglia.18 Together, these observations raise the possibility that heterotypic trans interactions of NRP1 with endothelial molecules such as VEGFR2 contribute to brain angiogenesis, as proposed for tumor angiogenesis.
Here, we have used Cre/Lox recombination to show that loss of NRP1 expression from proangiogenic macrophages or neural progenitors does not perturb brain vascularization, ruling out an essential role for nonendothelial NRP1 in this process. In contrast, targeting of conditional Nrp1-null alleles in endothelial cells with constitutively active or tamoxifen-inducible CRE recombinase confirmed a cell autonomous role for NRP1 in endothelial cells and further demonstrated that NRP1 confers a selective advantage to endothelial cells competing for the tip cell position in vessel sprouts. Thus, recombination-resistant cells in endothelial Nrp1-null mutants preferentially adopted the tip cell position and partially rescued the angiogenic defects observed in full Nrp1 knockouts. We conclude that NRP1 promotes endothelial tip cell function during sprouting angiogenesis.
Materials and Methods
Mouse strains
All animal procedures were performed in accordance with institutional and UK Home Office guidelines. Mice were mated in the evening, and the morning of vaginal plug formation was counted as embryonic day (E) 0.5. To delete NRP1 in selected cell types, we mated conditional Nrp1-null (floxed) mice (Nrp1fl/fl)22 to mice carrying one complete Nrp1 allele (Nrp1+/–)23 together with one of the following transgenes: Tie2-Cre,24 Nes-Cre,25 Csf1r-iCre,26 and Pdgfb-iCreER-Egfp27 with codon-improved Cre, or Cdh5(PAC)-CreER (referred to briefly as Cdh5-CreER).28 For tamoxifen-induced, Cre-mediated recombination of floxed target genes, 4-hydroxytamoxifen (4-OHT, Sigma, St. Louis, MO) was dissolved in absolute ethanol at 50 mg/mL, diluted in peanut oil to 5 mg/mL, and then administered via intraperitoneal injection into pregnant dams on day E9.5 (1 mg for Pdgfb-iCreER-Egfp, 2 mg for Cdh5-CreER). Mice containing the floxed Rosa26Yfp reporter have been described previously.29 Genotyping protocols can be supplied on request.
Immunolabeling and imaging
The following primary antibodies were used: rat anti-PECAM (BD Biosciences, Oxford, UK), rabbit anti-GFP (MBL International, Woburn, MA), rabbit anti-IBA1 (Wako Chemicals, Richmond, VA), rat anti-F4/80 (ABD Serotec, Kidlington, UK), goat anti-rat NRP1, anti-VEGFR2, and anti-TIE2 (R&D Systems, Minneapolis, MN). Secondary antibodies used included Alexa-conjugated goat anti-rat or anti-rabbit IgG (Life Technologies, Carlsbad, CA), and Cy3- or Alexa647-conjugated rabbit anti-goat Fab fragments (Jackson ImmunoResearch, West Grove, PA). In some experiments, anti-PECAM was detected by horseradish peroxidase-conjugated rabbit anti-rat IgG (DAKO UK, Ely, UK). To detect blood vessels, we also used biotinylated isolectin B4 (IB4; Sigma) followed by Alexa-conjugated streptavidin (Life Technologies). Samples were imaged with the LSM510 or LSM710 laser scanning confocal microscopes (Zeiss, Jena, Germany) or an MZ16 stereomicroscope (Leica, Wetzlar, Germany). Images were processed with Adobe Photoshop CS4 (Adobe Inc.). Three-dimensional rendering of high-resolution confocal z-stacks was carried out with Imaris (BitPlane, South Windsor, CT).
Fluorescence-activated cell sorting (FACS)
E11.5 whole embryos were mechanically homogenized in ice-cold RPMI1640 medium (Life Technologies) containing 5% fetal calf serum, 2.38g/L HEPES and 1.5g/L sodium hydrogen carbonate and passed through a 70-μm filter. The resulting cell suspensions were incubated with Fc block (Becton Dickinson, Oxford, UK) to prevent nonspecific binding of antibodies and then stained with APC-conjugated antibodies specific for PECAM (BD Pharmingen) to label endothelial cells and with DAPI to identify dying or dead cells. Labeled cells were analyzed with a BD LSR II flow cytometer (BD Biosciences). Samples from unstained control and Tie2-Cre–negative embryos were used to identify appropriate fluorescence voltage and gate parameters.
Quantification and statistical analysis
Vascular branchpoints at E12.5 were counted in three randomly chosen 0.25-mm2 regions per PECAM-labeled hindbrain and then averaged to obtain a value for each hindbrain. Recombination-resistant endothelial tip and stalk cells or NRP1-positive macrophages were counted in two randomly chosen 0.85-mm2 or 0.07-mm2 regions, respectively, for each YFP/NRP1/IB4-triple-labeled E11.5 hindbrain and then averaged to obtain a value for each hindbrain. For all experiments, we calculated the mean of 3 to 12 independent samples. Error bars represent the standard deviation of the mean. To determine whether two datasets were significantly different, we calculated the P value by performing a two-tailed unpaired Student’s t-test; to compare more than two datasets, we additionally performed a one-way analysis of variance followed by Tukey’s post-hoc test; a P value < .05 was considered significant. To determine whether the number of tip cells with a specific marker correlated with the number of vessel branchpoints, we determined the coefficient of determination, r2. All statistical analyses were performed with Prism 5 (GraphPad Software, La Jolla, CA).
Results
NRP1 is coexpressed by endothelial and nonendothelial cells during organ vascularization and is enriched in endothelial tip cells
Vessel growth in the embryonic central nervous system is severely compromised by loss of NRP1.14,17,22 Using a NRP1 antibody validated to stain embryonic wild-type, but not Nrp1-null, hindbrains,18 we found that NRP1 was coexpressed by endothelial cells and cells in their environment during the formation of the subventricular vascular plexus (SVP) that supplies the neural progenitor zone (Figure 1). Within the brain vascular endothelium, which is positive for isolectin B4 (IB4),18 NRP1 was expressed by both stalk and tip cells (Figure 1A,A′,B,B′; wavy arrows and arrowheads, respectively). NRP1 was particularly prominent on tip cell filopodia, similar to VEGFR2 (Figure 1A,F; arrowheads). In addition, NRP1 protein was expressed on the neural progenitors of the ventricular zone, which attract growing vessels into the brain (Figure 1A′-C; note tip cell filopodia protruding into the neural progenitor layer, indicated by solid arrowheads in Figure 1C). Finally, NRP1 was present on hindbrain tissue macrophages (Figure 1D-E), which were recognized by co-labeling for IB4 and IBA1.30,31 We have previously reported that these cells promote the anastomosis of nascent vessel sprouts in the developing brain.18 NRP1 expression in macrophages was higher at E11.5 (Figure 1E; solid arrow) than at E10.5, when some macrophages appeared poorly ramified and NRP1 negative (Figure 1A; solid arrow). At E11.5, NRP1 appeared enriched on the IBA1-positive macrophage processes that extended towards endothelial tip cells (Figure 1D′-E; clear arrows). Together, these expression patterns are consistent with the possibility that NRP1 regulates brain angiogenesis by acting on endothelial VEGFR2 either homotypically or heterotypically (Figure 1G).
NRP1 expression during hindbrain vascularization in the mouse. (A-F) Whole-mount immunofluorescence labeling of the mouse embryo hindbrain; scale bar (A-D and F) 50 μm. (A) Maximal projection (xy) of a confocal z stack through the E10.5 subventricular zone shows NRP1 on IB4-positive endothelial stalk and tip cells (wavy arrow and arrowhead, respectively; yellow indicates double labeling). NRP1 is also prominent on tip-cell filopodia (IB4 low) and neural progenitors (IB4 negative). (A′) Virtual transverse (xz) section through the z stack in (A) at the level indicated with a stippled line. (B,B′) Single NRP1 channel of (A,A′). Some IB4-positive tissue macrophages (arrow in A) are poorly ramified at E10.5 and do not obviously express NRP1 (compare A with B). (C) Single xy scan of the confocal z stack projection in (B) at the level indicated with a stippled line in (B'); clusters of tip cell filopodia protrude into the neural progenitor layer (arrowheads). (D-E) Confocal z stack through the E11.5 subventricular zone shows that IB4-positive endothelial cells and IBA1/IB4-positive ramified tissue macrophages express NRP1; purple indicates co-labeling with NRP1 and IB4. The boxed area in (D) is indicated at higher magnification in (D′) and as the single NRP1 channel in (E). NRP1 is high on filopodia-studded tip cells and on IBA1-enriched macrophage processes (arrowheads and clear arrows, respectively). NRP1 appears lower in neural progenitors at E11.5 (D) compared with E10.5 (A). (F) Z stack through the subventricular zone at E11.5 shows VEGFR2 expression on IB4-positive endothelial stalk and tip cells (wavy arrow and arrowhead, respectively; yellow indicates double labeling). VEGFR2 is high on filopodia but not obviously expressed by tissue macrophages or neural progenitors. (G) Schematic representation of NRP1 and VEGFR2 distribution and hypothetical interactions in hindbrain cell types during angiogenesis. NRP1 (light green) is co-expressed with VEGFR2 (dark green) on endothelial stalk and tip cells (red), which is a prerequisite for homotypic interactions. NRP1 is also expressed by neural progenitors (orange) and tissue macrophages (Mφ, blue), and therefore heterotypically and in trans relative to VEGFR2 in endothelial cells.
NRP1 expression during hindbrain vascularization in the mouse. (A-F) Whole-mount immunofluorescence labeling of the mouse embryo hindbrain; scale bar (A-D and F) 50 μm. (A) Maximal projection (xy) of a confocal z stack through the E10.5 subventricular zone shows NRP1 on IB4-positive endothelial stalk and tip cells (wavy arrow and arrowhead, respectively; yellow indicates double labeling). NRP1 is also prominent on tip-cell filopodia (IB4 low) and neural progenitors (IB4 negative). (A′) Virtual transverse (xz) section through the z stack in (A) at the level indicated with a stippled line. (B,B′) Single NRP1 channel of (A,A′). Some IB4-positive tissue macrophages (arrow in A) are poorly ramified at E10.5 and do not obviously express NRP1 (compare A with B). (C) Single xy scan of the confocal z stack projection in (B) at the level indicated with a stippled line in (B'); clusters of tip cell filopodia protrude into the neural progenitor layer (arrowheads). (D-E) Confocal z stack through the E11.5 subventricular zone shows that IB4-positive endothelial cells and IBA1/IB4-positive ramified tissue macrophages express NRP1; purple indicates co-labeling with NRP1 and IB4. The boxed area in (D) is indicated at higher magnification in (D′) and as the single NRP1 channel in (E). NRP1 is high on filopodia-studded tip cells and on IBA1-enriched macrophage processes (arrowheads and clear arrows, respectively). NRP1 appears lower in neural progenitors at E11.5 (D) compared with E10.5 (A). (F) Z stack through the subventricular zone at E11.5 shows VEGFR2 expression on IB4-positive endothelial stalk and tip cells (wavy arrow and arrowhead, respectively; yellow indicates double labeling). VEGFR2 is high on filopodia but not obviously expressed by tissue macrophages or neural progenitors. (G) Schematic representation of NRP1 and VEGFR2 distribution and hypothetical interactions in hindbrain cell types during angiogenesis. NRP1 (light green) is co-expressed with VEGFR2 (dark green) on endothelial stalk and tip cells (red), which is a prerequisite for homotypic interactions. NRP1 is also expressed by neural progenitors (orange) and tissue macrophages (Mφ, blue), and therefore heterotypically and in trans relative to VEGFR2 in endothelial cells.
Reduced severity of vascular defects in Tie2-Cre–targeted Nrp1 mutants compared with full Nrp1 knockouts
To clarify the contribution of endothelial versus nonendothelial NRP1 to brain angiogenesis, we compared vascular phenotypes in cell-type–specific Nrp1-null mutants, taking advantage of a previously published Nrp1 allele that can be inactivated by Cre/Lox recombination.22 To delete NRP1 in endothelial cells, we used a Cre transgene driven by the promoter for the gene encoding the angiopoietin receptor TIE2 (Tie2-Cre), because this gene-targeting strategy was previously shown to cause vascular defects in the brain.22 Because it was reported that Nrp1 targeting was more efficient with one conditional Nrp1-null allele on a heterozygous, constitutive Nrp1-null background (Nrp1fl/–) than with two conditional Nrp1-null alleles (Nrp1fl/fl),22 we also adopted this strategy to maximize possible phenotypes.
Immunohistochemistry for the endothelial marker PECAM showed a mild reduction in vessel branching in the subventricular zone of Tie2-Cre;Nrp1fl/+ and Nrp1fl/– heterozygous mutants compared with littermate Nrp1fl/+ controls (Figure 2A,B and data not shown). As expected, branching was more severely perturbed in Tie2-Cre;Nrp1fl/– mutants, which lack NRP1 expression in endothelial cells from both alleles (Figure 2C). Although vascular morphology in heterozygous hindbrains appeared grossly normal, except for a small reduction in vessel density, only few vessels had formed bridges in homozygous Tie2-Cre;Nrp1fl/– mutants (Figure 2C; clear arrow); instead, many vessels terminated in blind-ended tufts (Figure 2C; clear arrowheads). Unexpectedly, the vascular defect of hindbrains from complete Nrp1-null embryos was more severe than that of Tie2-Cre;Nrp1fl/– hindbrains; thus, most vessels terminated in tufts, and the tufts were often larger in Nrp1–/– hindbrains (compare vascular structures indicated with clear arrowheads in Figure 2C,D). Moreover, neighboring vessels failed to interconnect in the subventricular zone of Nrp1–/– hindbrains, and the few lateral connections that formed were located beneath the subventricular zone and therefore appeared out of focus in the whole-mount view (Figure 2D; arrows).
Vascular defects in Tie2-Cre conditional and full Nrp1-null mutants. (A-D) PECAM immunohistochemistry of E12.5 SVP vessels of the indicated genotypes. Clear arrowheads indicate examples of vascular tufts; clear arrows and solid arrows indicate examples of vascular interconnections in the SVP versus deeper brain layers, respectively. Scale bar represents 100 μm. (E) Quantitation of SVP branchpoints at E12.5; error bars represent SD; asterisks indicate P values; **P < .001; ***P < .0001.
Vascular defects in Tie2-Cre conditional and full Nrp1-null mutants. (A-D) PECAM immunohistochemistry of E12.5 SVP vessels of the indicated genotypes. Clear arrowheads indicate examples of vascular tufts; clear arrows and solid arrows indicate examples of vascular interconnections in the SVP versus deeper brain layers, respectively. Scale bar represents 100 μm. (E) Quantitation of SVP branchpoints at E12.5; error bars represent SD; asterisks indicate P values; **P < .001; ***P < .0001.
Quantitation of vascular branchpoints in the SVP confirmed a similar but small reduction in heterozygous Nrp1fl/– and Tie2-Cre;Nrp1fl/+mutants and a greater reduction in vessel branching in homozygous Tie2-Cre;Nrp1fl/– mutants compared with littermates with normal NRP1 expression (Figure 2E; Nrp1fl/+ controls 88.8 ± 7.3, n = 12; pooled Nrp1fl/– and Tie2-Cre;Nrp1fl/+ heterozygous mutants 71.2 ± 4.2, n = 16; Tie2-Cre;Nrp1fl/– homozygous mutants 33.2 ± 12.6, n = 5; P = .01 wildtype vs heterozygous mutants; P < .001 wildtype vs homozygous or heterozygous vs homozygous mutants). This analysis further demonstrated that the branching defect was significantly more severe in complete Nrp1-knockouts compared with endothelial Tie2-Cre;Nrp1fl/– mutants (Figure 2E; Tie2-Cre;Nrp1fl/– 33.2 ± 12.6, n = 5, vs Nrp1–/– 3.1 ± 2.0, n = 8; P = .01). We therefore investigated the possibility that NRP1 expression by cell types interacting with endothelial cells in trans contributes to hindbrain angiogenesis by ablating NRP1 in tissue macrophages and neural progenitors, respectively.
NRP1 expression by macrophages is not essential for brain vascularization
To define the contribution of macrophage NRP1 to hindbrain angiogenesis, we could not take advantage of the LysmCre line previously used by others to study the function of myeloid genes, because this CRE allele fails to target tissue macrophages in the mouse embryo hindbrain.18 We therefore expressed Cre under control of the promoter for the CSF1 receptor gene (Csf1r-iCre), a strategy previously shown to target monocyte-derived macrophages.26 To confirm that Csf1r-iCre also deletes in the yolk sac–derived tissue macrophages during brain vascularization, we introduced this transgene into the floxed Rosa26Yfp reporter knockin mouse, which produces yellow fluorescent protein (YFP) in cells expressing Cre recombinase.29 Immunolabeling of hindbrains carrying the Csf1r-iCre transgene demonstrated efficient activation of Rosa26Yfp in IB4-positive tissue macrophages at E10.5 and established that 98% of F4/80- and IB4-positive macrophages in the E11.5 hindbrain were YFP-positive (Figure 3A-A′′′ and data not shown). Because NRP1 expression was low on macrophages at E10.5 and partially obscured by strong neural progenitor NRP1 expression at that stage (Figures 1 and 3), we used immunolabelling at E11.5 to determine the efficiency of NRP1 knockdown in tissue macrophages of Csf1r-iCre;Nrp1fl/– hindbrains (Figure 3B-C). The quantitative analysis of immunolabeled hindbrains showed that 90% ± 5% of tissue macrophages normally expressed NRP1 in Nrp1fl/– controls, whereas tissue macrophages expressing NRP1 at detectable levels could not be identified in Csf1riCre;Nrp1fl/– hindbrains (n = 3; Figure 3C′; asterisks).
NRP1 expression by tissue macrophages is not essential for brain vascularization. (A) A E10.5 hindbrain with a constitutively active Csf1r-iCre transgene and the Rosa26Yfp reporter was triple labeled for YFP (green), NRP1 (red), and IB4 (blue), shown together (A) and as single channels (A′-A′′′). The solid arrow indicates a tissue macrophage, the clear arrow a NRP1-positive macrophage process; the solid arrowhead indicates a tip cell. Scale bar represents 50 μm. (B,C) Double labeling of E11.5 control Nrp1fl/– and mutant Csf1r-iCre;Nrp1fl/– hindbrains for NRP1 (red) and IB4 (blue) shown together (B,C) and as single NRP1 channels (B′,C′). A NRP1-positive tissue macrophage is indicated with an arrow in (B,B′); NRP1-negative tissue macrophages are indicated with asterisks in (C,C′). Scale bar represents 25 μm. (D,E) PECAM immunohistochemistry of E12.5 littermate hindbrains of the indicated genotypes; scale bar represents 100 μm. (F) Quantitation of SVP branchpoints at E12.5; error bars represent SD; n.s., not significant.
NRP1 expression by tissue macrophages is not essential for brain vascularization. (A) A E10.5 hindbrain with a constitutively active Csf1r-iCre transgene and the Rosa26Yfp reporter was triple labeled for YFP (green), NRP1 (red), and IB4 (blue), shown together (A) and as single channels (A′-A′′′). The solid arrow indicates a tissue macrophage, the clear arrow a NRP1-positive macrophage process; the solid arrowhead indicates a tip cell. Scale bar represents 50 μm. (B,C) Double labeling of E11.5 control Nrp1fl/– and mutant Csf1r-iCre;Nrp1fl/– hindbrains for NRP1 (red) and IB4 (blue) shown together (B,C) and as single NRP1 channels (B′,C′). A NRP1-positive tissue macrophage is indicated with an arrow in (B,B′); NRP1-negative tissue macrophages are indicated with asterisks in (C,C′). Scale bar represents 25 μm. (D,E) PECAM immunohistochemistry of E12.5 littermate hindbrains of the indicated genotypes; scale bar represents 100 μm. (F) Quantitation of SVP branchpoints at E12.5; error bars represent SD; n.s., not significant.
Immunohistochemistry for PECAM at E12.5 showed that the brain vasculature had developed similarly in Csf1r-iCre–targeted Nrp1fl/– mutants and controls (Figure 3D,E; controls were Nrp1fl/– littermates to account for a small reduction in vessel branching in heterozygous hindbrains, as shown in Figure 2). Quantitation of the number of SVP branchpoints confirmed similar hindbrain vascularization in homozygous mutants and controls (Figure 3F; Nrp1fl/+ 84.9 ± 3.0, n = 6; Nrp1fl/– 80.0 ± 4.6, n = 6, vs Csf1r-iCre;Nrp1fl/– 77.4 ± 8.3, n = 5). These observations show that NRP1, despite being a good marker for the macrophages that promote vascular anastomosis,18,32 is not essential for the proangiogenic function of these cells during development.
NRP1 expression by neural progenitors is not essential for brain vascularization
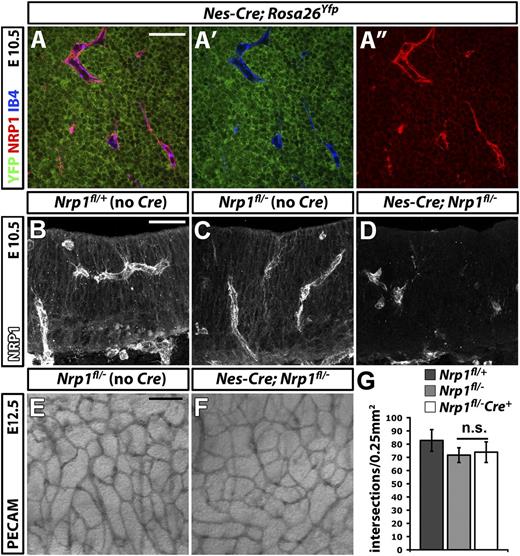
To delete NRP1 in neural progenitors, we expressed CRE under the control of the nestin promoter (Figure 4). For these experiments, we used a transgenic mouse line that targets neural progenitors from E8.5 onward (Nes-Cre),25 ie before the onset of brain vascularization at E9.5.18 Immunolabeling of E10.5 hindbrains carrying the Nes-Cre transgene demonstrated efficient activation of Rosa26Yfp in neural progenitors that expressed NRP1 (Figure 4A-A″). Moreover, NRP1 was effectively knocked down in neural progenitors in Nes-Cre;Nrp1fl/– mutants, demonstrating that Nes-Cre is a suitable tool to target NRP1 expression in these cells (Figure 4B,D). The brain vasculature of Nes-Cre;Nrp1fl/– mutants appeared similar to that of Nrp1fl/– littermate controls (Figure 4E,F). Moreover, the number of SVP branchpoints was comparable in both genotypes (Figure 4G; Nrp1fl/+ 82.8 ± 8.2, n = 3; Nrp1fl/– 71.2 ± 5.6, n = 5, vs Nes-Cre;Nrp1fl/– 73.9 ± 7.8, n = 8). Accordingly, NRP1 expression by neural progenitors does not make an essential contribution to NRP1-mediated brain vascularization. In summary, heterotypic NRP1 expression by neural progenitors or tissue macrophages is not essential for brain vascularization.
NRP1 expression by neural progenitors is not essential for brain vascularization. (A-A″) Single xy scan through an E10.5 hindbrain carrying the constitutively active Nes-Cre transgene and the Rosa26Yfp reporter, labeled for YFP (green), NRP1 (red), and IB4 (blue), all shown together in (A) or as double YFP/IB4 (A′) and single NRP1 (A″) channels. Scale bar represents 50 μm. (B-D) Immunofluorescent staining for NRP1 of 20-μm thin, frozen sections from E10.5 control Nrp1fl/+, Nrp1fl/–, and mutant Nes-Cre;Nrp1fl/– hindbrains. Scale bar represents 50 μm. (E,F) PECAM immunohistochemistry of E12.5 littermate hindbrains of the indicated genotypes; scale bar represents 100 μm. (G) Quantitation of SVP branchpoints at E12.5; error bars represent SD; n.s., not significant.
NRP1 expression by neural progenitors is not essential for brain vascularization. (A-A″) Single xy scan through an E10.5 hindbrain carrying the constitutively active Nes-Cre transgene and the Rosa26Yfp reporter, labeled for YFP (green), NRP1 (red), and IB4 (blue), all shown together in (A) or as double YFP/IB4 (A′) and single NRP1 (A″) channels. Scale bar represents 50 μm. (B-D) Immunofluorescent staining for NRP1 of 20-μm thin, frozen sections from E10.5 control Nrp1fl/+, Nrp1fl/–, and mutant Nes-Cre;Nrp1fl/– hindbrains. Scale bar represents 50 μm. (E,F) PECAM immunohistochemistry of E12.5 littermate hindbrains of the indicated genotypes; scale bar represents 100 μm. (G) Quantitation of SVP branchpoints at E12.5; error bars represent SD; n.s., not significant.
Tie2-Cre effectively ablates NRP1 in macrophages and endothelial stalk but not in tip cells
The experiments described here showed that NRP1 expression by nonendothelial cells is dispensable for hindbrain angiogenesis (Figures 3 and 4). However, the Tie2-Cre–mediated endothelial knockdown of NRP1 did not recapitulate the severity of vascular defects in the full Nrp1 knockout (Figure 2). We therefore asked whether Tie2-Cre is a good tool to target the brain vasculature. Immunolabeling of E11.25 hindbrains confirmed that neural progenitors retained NRP1 in Tie2-Cre–targeted Nrp1 mutants and their littermate controls lacking Tie2-Cre (Figure 5). Unexpectedly, yolk sac–derived tissue macrophages lacked NRP1 expression in mutant hindbrains expressing Cre, even though they expressed NRP1 in control hindbrains lacking Cre (compare Figure 5A,B with 5 C,D; arrows in Figure 5B,B′ indicate IB4/NRP1–double-positive tissue macrophages in control hindbrains; asterisks in Figure 5D′ indicate the position of NRP1-negative tissue macrophages that are recognized by IB4 staining and indicated with clear arrows in Figure 5D). Because the macrophage-selective deletion of Nrp1 does not impair brain angiogenesis (Figure 3), the vascular phenotype of Tie2-Cre–targeted Nrp1 mutants could not be attributed to NRP1 function in tissue macrophages.
Tie2-Cre;Nrp1fl/– mutants contain tip cells that retain NRP1 expression, even though Tie2-Cre effectively activates the Rosa26Yfp reporter in tissue macrophages as well as endothelial tip and stalk cells. (A-D) NRP1 (red) and IB4 (green) immunofluorescence staining of littermate E11.25 hindbrains lacking Cre or expressing a constitutively active Tie2-Cre transgene on an Nrp1fl/– background; single NRP1 channels are shown below each panel (A′-D′). (B and D) Higher magnifications of the boxed areas in (A and C). Arrowheads indicate examples of tip cells, arrows show examples of tissue macrophages expressing NRP1. Clear arrows in (D) and asterisks in (D′) indicate the position of macrophages lacking NRP1, and curved arrows indicate endothelial stalk cells lacking NRP1. Scale bar represents 100 μm for (A,A′,C,C′). (E) An E11.5 hindbrain carrying a constitutively active Tie2-Cre transgene and the Rosa26Yfp reporter was triple labeled for NRP1 (red), YFP (green), and IB4 (blue); single channels are shown in (E′-E′′′). Scale bar represents 50 μm.
Tie2-Cre;Nrp1fl/– mutants contain tip cells that retain NRP1 expression, even though Tie2-Cre effectively activates the Rosa26Yfp reporter in tissue macrophages as well as endothelial tip and stalk cells. (A-D) NRP1 (red) and IB4 (green) immunofluorescence staining of littermate E11.25 hindbrains lacking Cre or expressing a constitutively active Tie2-Cre transgene on an Nrp1fl/– background; single NRP1 channels are shown below each panel (A′-D′). (B and D) Higher magnifications of the boxed areas in (A and C). Arrowheads indicate examples of tip cells, arrows show examples of tissue macrophages expressing NRP1. Clear arrows in (D) and asterisks in (D′) indicate the position of macrophages lacking NRP1, and curved arrows indicate endothelial stalk cells lacking NRP1. Scale bar represents 100 μm for (A,A′,C,C′). (E) An E11.5 hindbrain carrying a constitutively active Tie2-Cre transgene and the Rosa26Yfp reporter was triple labeled for NRP1 (red), YFP (green), and IB4 (blue); single channels are shown in (E′-E′′′). Scale bar represents 50 μm.
We therefore examined next the deletion of NRP1 within the vascular endothelium. Surprisingly, the Tie2-Cre–mediated knockdown of NRP1 in endothelium was incomplete, with mutants containing NRP1-positive, filopodia-studded tip cells at the front of vessel sprouts (compare Figure 5A,B with 5C,D; solid arrowheads indicate examples of tip cells in Figure 5 B-B′ and 5D-D′). Yet, intervening vessel segments, composed of stalk cells, appeared NRP1 negative, as expected (Figure 5D; curved arrows). The presence of a small number of NRP1-positive tip cells leading new vessel sprouts suggested that Tie2-Cre–mediated targeting had spared NRP1 in a subset of endothelial cells.
Two alternative hypotheses may explain why tip cells were spared from NRP1 loss in Tie2-Cre;Nrp1fl/– mutants. One possibility is that Tie2-Cre targets stalk, but not tip cells. The alternative possibility is that a subset of recombination-resistant endothelial cells preferentially assumes a tip rather than stalk cell position, because NRP1 confers a selective advantage to cells competing for the tip cell position. To distinguish these possibilities, we next studied hindbrains carrying the Rosa26Yfp reporter.
Tie2-Cre effectively targets tissue macrophages, endothelial tip, and endothelial stalk cells
To investigate whether Tie2-Cre preferentially targets stalk over tip cells, we immunolabeled E11.5 hindbrains carrying the Rosa26Yfp reporter. We observed YFP expression in IB4/NRP1-positive tissue macrophages as well as endothelial tip and endothelial stalk cells (Figure 5E-E′′′ and Figure 6A,A′,B; solid arrows indicate examples of tissue macrophages, solid arrowheads examples of endothelial tip cells, and wavy arrows examples of endothelial stalk cells). Co-targeting of endothelial cells and tissue macrophages may reflect Tie2 expression in a shared progenitor or active expression of Tie2 in both cell types during brain angiogenesis. Consistent with the latter, but not excluding the former possibility, the TIE2 protein localized to F4/80-positive tissue macrophages, in addition to endothelial tip and stalk cells (supplemental Figure 1A-A′′′; the arrowhead indicates a tip cell, the wavy arrow a vessel segment composed of stalk cells; arrows highlight tissue macrophages). Tie2-Cre activity therefore reflects the expression of endogenous TIE2 in tissue macrophages and endothelial cells during brain angiogenesis. In addition, targeted cells may be derived from a shared, TIE2-positive precursor.
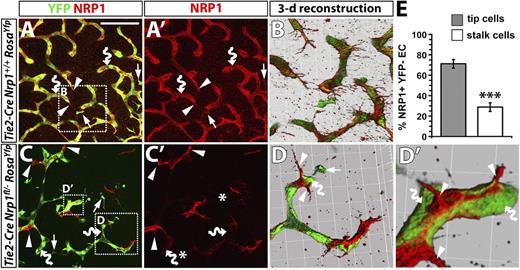
NRP1 expression confers a selective advantage to endothelial cells competing for the tip cell position. (A-D) Immunofluorescence staining of littermate hindbrains of the indicated genotypes; YFP and NRP1 labeling are shown in (A,C), NRP1 only in (A′,C′). Scale bar represents 100 μm. Three-dimensional reconstructions of the boxed areas in (A,C) shown in (B,D,D′). Examples of tissue macrophages, endothelial tip cells, and endothelial stalk cells are indicated with arrows, arrowheads, and curved arrows, respectively; note that some vessels leave the plane of section, and the vessel therefore appears blunt-ended, terminating in a stalk cell. Asterisks in (C′) indicate the position of macrophages lacking NRP1. (E) Percentage of NRP1+ YFP– endothelial cells in the tip versus stalk cell position in Tie2-Cre;Nrp1fl/–;RosaYfp mutant hindbrains.
NRP1 expression confers a selective advantage to endothelial cells competing for the tip cell position. (A-D) Immunofluorescence staining of littermate hindbrains of the indicated genotypes; YFP and NRP1 labeling are shown in (A,C), NRP1 only in (A′,C′). Scale bar represents 100 μm. Three-dimensional reconstructions of the boxed areas in (A,C) shown in (B,D,D′). Examples of tissue macrophages, endothelial tip cells, and endothelial stalk cells are indicated with arrows, arrowheads, and curved arrows, respectively; note that some vessels leave the plane of section, and the vessel therefore appears blunt-ended, terminating in a stalk cell. Asterisks in (C′) indicate the position of macrophages lacking NRP1. (E) Percentage of NRP1+ YFP– endothelial cells in the tip versus stalk cell position in Tie2-Cre;Nrp1fl/–;RosaYfp mutant hindbrains.
The finding that Tie2-Cre is not selective for tip or stalk cells agrees with previous studies in the retina and embryoid bodies, which demonstrated that tip and stalk cells are not a lineage-specified subtype of endothelial cells.7 Instead, a proportion of endothelial cells is temporarily and dynamically singled out from the endothelial cell population after exposure to VEGF-A gradients and upregulation of DLL4 to serve as tip cells that guide vessel sprouting.5,6,33 Because tip cells are not a different lineage than stalk cells, and Tie2-Cre can in principle recombine in both cell populations, the NRP1-positive tip cells in Tie2-Cre;Nrp1fl/– mutants have therefore more likely arisen from endothelial cells that did not undergo recombination for unknown reasons and, because of their retention of NRP1 expression, adopted the leading position in vessel sprouts.
Endothelial cells resisting Tie2-Cre recombination adopt tip cell positions and promote vessel branching
To examine whether NRP1-positive tip cells in Tie2-Cre;Nrp1fl/– hindbrains arose from recombination-resistant endothelial cells, we introduced the Rosa26Yfp reporter into this conditional Nrp1-null strain and compared YFP expression in Tie2-Cre;Nrp1+/+ versus Tie2-Cre;Nrp1fl/– littermates (Figure 6). In agreement with our previous observations, control hindbrains expressed YFP in tissue macrophages as well as in tip and stalk cells (Figure 6A,A′). Accordingly, 3-dimensional reconstructions of confocal z-stacks through the subventricular zone showed YFP-positive stalk cells with NRP1 surface labeling (Figure 6B). In Tie2-Cre;Nrp1fl/– mutants, stalk cells deficient in NRP1 were YFP-positive, as expected, whereas tip cells that retained NRP1 lacked YFP expression (Figure 6C,C′). Consequently, 3-dimensional reconstructions showed vessel branches in Tie2-Cre;Nrp1fl/– that were composed of YFP-positive stalk cells led by NRP1-positive tip cells (Figure 6D,D′). Quantitation demonstrated that NRP1-positive endothelial cells in Tie2-Cre;Nrp1fl/– mutants preferentially adopted tip rather than stalk cell positions (Figure 6E).
We next asked whether the large number of recombination-resistant endothelial cells in the prominent tip cell position of Tie2-Cre;Nrp1fl/– vessels simply reflected generally less efficient recombination across the vasculature in mutants compared with controls. We used FACS to separate cell suspensions from mutants and littermate controls on a Rosa26Yfp background into PECAM-positive and PECAM-negative pools and further gated these cells into YFP-positive and YFP-negative subpopulations (Figure 7A,C). The quantitative analysis of the sorted populations demonstrated a similar ratio of endothelial to nonendothelial cells in mutants and controls (Figure 7D). Tie2-Cre targeting of Nrp1 therefore did not alter the total number of endothelial cells. Furthermore, the proportion of endothelial cells expressing or lacking YFP was similar in both genotypes (Figure 7E), with a high proportion being gene-targeted and a low proportion not targeted (controls: 93.6 ± 1.5% targeted and 6.4 ± 1.5% nontargeted cells, n = 6; Tie2-Cre;Nrp1fl/–: 92.8 ± 0.3% targeted and 7.2 ± 0.3% nontargeted cells, n = 3). Accordingly, Tie2-Cre–mediated recombination occurred with similar efficiency in endothelial cells of Tie2-Cre;Nrp1fl/– mutants and controls.
The number of NRP1-positive tip cells in blood vessels with mosaic expression of NRP1 is a good predictor of branching frequency. (A-C) Live cells from the indicated genotypes were FACS-sorted with a PECAM-APC antibody (y axis) and for YFP fluorescence (x axis); (A) Cells from a control embryo lacking YFP and not stained for PECAM; this FACS profile was used to determine gating parameters to identify YFP-positive, PECAM-stained cells in (B,C). (D) Percentage of total PECAM-positive (Q1 + Q2 quadrants) and PECAM-negative (Q3 + Q4 quadrants) cells in (B and C). (E) Percentage of YFP-positive, PECAM-positive cells (Q2 quadrant) and YFP-negative, PECAM-positive cells (Q1 quadrant) in (B,C). (F,G) The number of vessel branchpoints in three littermate mutants lacking NRP1 in the Tie2-Cre lineage correlates inversely with the number of YFP-positive (F) and positively with the number of NRP1-positive tip cells (G). (H) Schematic representation of NRP1 localization in wild-type (top) and chimeric (bottom) vessel sprouts; NRP1 is present on both tip and stalk cells in wild-types (bright red), whereas NRP1 is high on mutant tip cells (bright red) and low on mutant stalk cells (faded red) of chimeric vessels.
The number of NRP1-positive tip cells in blood vessels with mosaic expression of NRP1 is a good predictor of branching frequency. (A-C) Live cells from the indicated genotypes were FACS-sorted with a PECAM-APC antibody (y axis) and for YFP fluorescence (x axis); (A) Cells from a control embryo lacking YFP and not stained for PECAM; this FACS profile was used to determine gating parameters to identify YFP-positive, PECAM-stained cells in (B,C). (D) Percentage of total PECAM-positive (Q1 + Q2 quadrants) and PECAM-negative (Q3 + Q4 quadrants) cells in (B and C). (E) Percentage of YFP-positive, PECAM-positive cells (Q2 quadrant) and YFP-negative, PECAM-positive cells (Q1 quadrant) in (B,C). (F,G) The number of vessel branchpoints in three littermate mutants lacking NRP1 in the Tie2-Cre lineage correlates inversely with the number of YFP-positive (F) and positively with the number of NRP1-positive tip cells (G). (H) Schematic representation of NRP1 localization in wild-type (top) and chimeric (bottom) vessel sprouts; NRP1 is present on both tip and stalk cells in wild-types (bright red), whereas NRP1 is high on mutant tip cells (bright red) and low on mutant stalk cells (faded red) of chimeric vessels.
Taking advantage of the small variation in recombination efficiency between individual embryos, we next asked whether the degree of gene targeting correlated with the phenotypic severity of the Tie2-Cre;Nrp1fl/– mutant vasculature. We observed that the number of YFP-positive, ie recombined tip cells inversely correlated with the number of vascular branchpoints in three Tie2-Cre;Nrp1fl/– littermate mutants (Figure 7F; r2 = 0.9124 for YFP-positive tip cells vs branchpoints; a value of 1.0 would indicate a perfect fit). Vice versa, the number of vessel sprouts with NRP1-retaining tip cells correlated positively with the number of branchpoints (Figure 7G; r2 = 0.8445 for NRP1-positive tip cells vs branchpoints). The number of NRP1-positive tip cells in an otherwise NRP1-negative vessel bed is therefore a good predictor of branching frequency. Consistent with the idea that NRP1-positive endothelial cells rescue vessel branching in mosaic blood vessel networks, the recombination-resistant (YFP-negative) tip cells in Tie2-Cre;Nrp1fl/– mutants retained high levels of VEGFR2 expression (supplemental Figure 2).
Tamoxifen-induced mosaic Nrp1 targeting confirms that NRP1-expressing endothelial cells have a selective advantage in adopting the tip cell position
To validate our findings in an alternative approach to Tie2-Cre targeting, we used a tamoxifen-inducible Cre transgene under the control of the endothelial Pdgfb promoter to ablate Nrp1 in endothelial cells (Pdgfb-iCreER-Egfp).27 Pdgfb is expressed at high levels in endothelial tip cells in the angiogenic retina,6,27 and Pdgfb-iCreER-Egfp is therefore particularly well suited to target these cells.27 To confirm that this transgene is also active in endothelium during hindbrain angiogenesis, we monitored the expression of eGFP, which is constitutively expressed from this transgene independently of tamoxifen-mediated CRE activation.27 This analysis confirmed that Pdgfb-iCreER-Egfp is suitable to target endothelial cells, but not tissue macrophages, in the developing hindbrain (supplemental Figure 3B-B′; arrows indicate macrophages that express NRP1 and IB4, but not GFP). In contrast to Tie2-Cre, Pdgfb-iCreER-Egfp therefore permits selective targeting of endothelial cells during brain angiogenesis.
Forty-eight hours after tamoxifen administration, vascular network complexity was reduced in Pdgfb-iCreER-Egfp;Nrp1fl/fl mutants compared with Nrp1fl/fl controls (supplemental Figure 3A,B; injection at E9.5, analysis at E11.5). This result again verified the endothelial requirement for NRP1 during hindbrain angiogenesis. However, as observed with Tie2-Cre, Pdgfb-iCreER-Egfp–mediated targeting of Nrp1 was incomplete, and the hindbrain endothelium was therefore mosaic with respect to NRP1 expression. Also similar to Tie2-Cre–mediated Nrp1 targeting, endothelial cells that had undergone CRE-mediated recombination in Pdgfb-iCreER-Egfp;Nrp1fl/fl mutant hindbrains were preferentially seen in stalk cell positions, whereas NRP1-retaining endothelial cells were predominantly found at the tip of vessel sprouts (curved arrows and arrowheads, respectively, in supplemental Figure 3B-B″). Thus, although Pdgfb-iCreER-Egfp targets endothelial tip cells,27 nontargeted endothelial cells that retained NRP1 expression had shuffled to tip cell positions 48 hours after tamoxifen administration. These observations are consistent with the previously reported dynamic competition of endothelial cells for the tip cell position in growing vessel sprouts.7
Finally, we performed mosaic deletion of Nrp1 in endothelial cells with another tamoxifen-inducible transgene that expresses Cre under the control of the VE-cadherin promoter (Cdh5-CreER).28 The Rosa26Yfp reporter showed that gene targeting was less efficient with this transgene than with Tie2-Cre or Pdgfb-iCreER-Egfp (eg, compare supplemental Figure 3B with supplemental Figure 4A,B). Nevertheless, we could detect YFP expression in both tip and stalk cells in control hindbrains (supplemental Figure 4A), whereas YFP-positive endothelial cells with low NRP1 levels remained in the stalk cell region in Cdh5-CreER;Nrp1fl/fl mutants (supplemental Figure 4B).
Taken together, gene targeting with constitutively active or tamoxifen-inducible endothelial Cre recombinase led to different degrees of mosaic NRP1 expression in the hindbrain endothelium, but in all cases, NRP1-retaining endothelial cells preferentially attained the tip cell position in vessel sprouts composed of NRP1-positive and NRP1-negative cells (Figure 7H).
Discussion
In addition to being prominent on vascular endothelial cells, NRP1 is expressed by tumor cells in adults and a wide range of cell types during development (eg, neurons, neural crest cells, macrophages).10,14,18,34-36 We have shown here that NRP1 expression by neural progenitors and macrophages is not required for brain angiogenesis, even though these cell types interact with angiogenic endothelium and can therefore present NRP1 in trans (Figures 1, 3, and 4). It is therefore likely that the ability of NRP1 to interact heterotypically with VEGFR2 in tissue culture models9 and homophilically in biochemical assays37 is not important for physiological angiogenesis, at least during brain vascular development. It remains to be investigated whether such interactions contribute to macrophage-vessel or tumor-vessel interactions in adults.
We have targeted endothelial cells with the Tie2-Cre transgene, which has been used to investigate the role of various angiogenesis-related genes in the vasculature. However, this Cre transgene is also active in the bone marrow–derived hematopoietic lineage,38 and adoptive transfer with wild-type bone marrow was therefore used in a tumor angiogenesis study to distinguish the role of HIF1A in monocytes and endothelium after Tie2-Cre targeting.39 Unexpectedly, we observed that Tie2-Cre also recombined floxed genes in tissue macrophages of the embryonic brain, which do not develop from bone marrow–derived hematopoietic cells or their embryonic equivalents, but instead differentiate in the yolk sac from hematopoietic progenitors without a monocyte intermediate.40,41 The remarkable efficiency with which Tie2-Cre targeted floxed genes in tissue macrophages may be because of TIE2 expression in their yolk sac progenitor, but it also agrees with the finding that differentiated tissue macrophages can themselves express TIE2 (supplemental Figure 1).18
Accordingly, Tie2-Cre–mediated targeting can establish the endothelial requirement of a gene under investigation only if that gene is not expressed in yolk sac–derived or bone marrow–derived myeloid cells. Should the gene be expressed by both macrophages and endothelial cells, Tie2-Cre targeting should be complemented by bone marrow reconstitution for adult studies39 or a second, macrophage- or endothelial-specific Cre-targeting approach to exclude a myeloid cell contribution to angiogenesis and establish a cell-autonomous endothelial function for the gene under investigation. Because the macrophage-restricted targeting of Nrp1 did not result in any vascular phenotype (Figure 3), we conclude that the vascular abnormalities observed in Tie2-Cre;Nrp1fl/– mutants reflect an essential role for NRP1 in endothelial cells. In agreement, Nrp1 targeting with an alternative endothelial Cre transgene that is not active in tissue macrophages confirmed an exclusively endothelial role for NRP1 in hindbrain angiogenesis (supplemental Figure 3).
Previously, the conditional ablation of Nrp1 expression with Tie2-Cre was reported to cause vessel defects similar to those of complete Nrp1-null mutants.22 We have confirmed this observation but demonstrated that vascular defects were far less severe than those of full knockouts (Figure 2). The reduced severity of the conditional mutants was not a result of compensation by NRP1-expressing macrophages or neural progenitors in the vascular environment, but was explained by inefficient Cre-Lox recombination in some endothelial cells. Thus, endothelial cells spared from recombination retained NRP1 and preferentially acquired the tip-cell position in angiogenic sprouts to partially restore tissue vascularization (Figures 5 and 6, supplemental Figure 3). We therefore extend the current model of angiogenesis by identifying NRP1 as an important gene promoting tip cell function. Furthermore, deletion of Nrp1 from endothelial cells did not alter the overall number of endothelial cells in the embryo (Figure 7), in agreement with a previous report showing that NRP1 does not control endothelial cell proliferation,16 a prominent feature of stalk cells.6 Because our data suggest that NRP1 expression in stalk cells is not essential for vessel sprouting or endothelial proliferation, stalk-cell expression may instead be indicative of later roles for NRP1 in endothelial survival, vascular maturation, or barrier maintenance.42-44
In our genetic analysis, a small proportion of endothelial cells consistently escaped Tie2-Cre– or Pdgfb-iCreER-Egfp–mediated recombination (Figures 6 and 7, supplemental Figure 3). Because the Rosa26Yfp reporter and eGFP expression demonstrated targeting of both tip and stalk cells in control hindbrains, the phenotype of these Nrp1 mutants is not likely caused by differential CRE activity in tip versus stalk cells. Instead, the inefficient targeting of floxed alleles in a small proportion of endothelial cells gave rise to mosaic endothelium in vivo, with individual endothelial cells succeeding in the competition for the tip cell position if they retained NRP1 expression. Thus, incomplete CRE recombination in endothelial cells provided in vivo data comparable with those derived from in vitro experiments with chimeric embryoid bodies, in which lineage-marked wild-type versus Vegfr2 haploinsufficient embryonic stem cells were differentiated into endothelial cells, and the wild-type cells with higher VEGFR2 levels had a selective advantage in becoming tip cells.7 NRP1 and VEGFR2 are therefore both important for the endothelial tip-cell phenotype, consistent with their ability to form complexes in vitro8 and their localization to tip-cell filopodia (Figure 1).
In conclusion, our analysis of cell-type–specific Nrp1 mouse mutants demonstrated a cell-autonomous role for NRP1 in endothelial tip cells to promote angiogenic vessel sprouting. Future work will need to determine the precise cellular function of NRP1 in tip cells. Thus, previous tissue culture studies suggested that endothelial NRP1 can promote VEGFR2 signaling,8,9 VEGFR2 endocytosis,45,46 VEGFR2 signal output,47 but also VEGFR2-independent VEGF-A signaling,43 integrin-mediated fibronectin assembly48 and matrix adhesion.49 However, which of these pathways involve NRP1 specifically during endothelial tip cell guidance and migration remains to be determined.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank David Ginty, Alex Kolodkin, Chenghua Gu, Ralf Adams, Hajime Fujisawa, and Weimin Zhong for providing mouse strains; Lena Claesson-Welsh for helpful discussions; Kathryn Davidson for help with genotyping; the staff of the Biological Resources Unit at the UCL Institute of Ophthalmology for help with mouse husbandry; the Imaging Facility of the UCL Institute of Ophthalmology for maintenance of the confocal microscopes; the Flow Cytometry Facility at the UCL Institute of Child Health; and Grazyna Galatowicz and Dawn Sim for help with FACS analysis.
This study was supported by British Heart Foundation grant PG/10/86/28622, MRC grant G0601093, and Wellcome Trust grant 095623/Z/11/Z (C.R.); by National Institutes of Health grants PO1 CA100324 and RO1 CA131270 (J.W.P.); and British Heart Foundation PhD studentship grant FS/10/54/28680 (A.P.).
Authorship
Contribution: A.F., J.M.V., and C.R. designed the research, performed the research, and analyzed the data; C.R. and A.F. wrote the paper; A.P and L.D. performed research and analyzed the data; J.W.P. and M.F. contributed essential reagents.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for J.M.V. is Department of Physiology, Anatomy and Genetics, University of Oxford, Oxford, United Kingdom.
Correspondence: Christiana Ruhrberg, UCL Institute of Ophthalmology, University College London, 11-43 Bath St, London EC1V 9EL, United Kingdom; e-mail: c.ruhrberg@ucl.ac.uk.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal