Key Points
We identify genes that are bound and regulated by Ikaros in pre-B cells.
Ikaros dosage drives the differentiation of cycling (Fr.C') to resting (Fr.D) pre-B cells.
Abstract
Ikaros family DNA-binding proteins are critical regulators of B-cell development. Because the current knowledge of Ikaros targets in B-cell progenitors is limited, we have identified genes that are bound and regulated by Ikaros in pre-B cells. To elucidate the role of Ikaros in B-cell lineage specification and differentiation, we analyzed the differential expression of Ikaros targets during the progression of multipotent to lymphoid-restricted progenitors, B- and T-cell lineage specification, and progression along the B-cell lineage. Ikaros targets accounted for one-half of all genes up-regulated during B-cell lineage specification in vivo, explaining the essential role of Ikaros in this process. Expression of the Ikaros paralogs Ikzf1 and Ikzf3 increases incrementally during B-cell progenitor differentiation, and, remarkably, inducible Ikaros expression in cycling pre-B cells was sufficient to drive transcriptional changes resembling the differentiation of cycling to resting pre-B cells in vivo. The data suggest that Ikaros transcription factor dosage drives the progression of progenitors along a predetermined lineage by regulating multiple targets in key pathways, including pre-B–cell receptor signaling, cell cycle progression, and lymphocyte receptor rearrangement. Our approach may be of general use to map the contribution of transcription factors to cell lineage commitment and differentiation.
Introduction
Ikaros DNA-binding proteins are important regulators of hematopoiesis1 and genetic deletion of Ikaros results in severe developmental disturbances, including delayed thymocyte differentiation and an early and complete block in B-cell development due to a failure of lymphoid-primed multipotent progenitors to progress toward common lymphoid progenitors (CLPs).1-3 Lymphoid malignancies arise in mice that are heterozygous for Ikzf1 (the gene encoding Ikaros)4 or that express Ikzf1 variants deficient in DNA binding.5,6 IKZF1 is frequently mutated or deleted in acute lymphoblastic leukemias, especially in cases with B-cell receptor (BCR)-ABL1 translocations, poor clinical outcome, and a high risk of relapse.7,8
B-cell development progresses through a sequence of developmental stages that are defined by the expression of specific marker genes and that require alterations between cell cycle progression and cell cycle arrest to facilitate the somatic rearrangement of BCR genes.9,10 Long-term self-renewing hematopoietic stem cells in the adult bone marrow differentiate into multipotent progenitor cells [also described as multilineage progenitors (MLP) or lymphoid-primed multipotent progenitor cells], which in turn form lineage-restricted progenitors, including CLPs and common myeloid progenitors. CLPs give rise to T-cell lineages and B-cell progenitors, which transit through stages designated as pre-pro-B or Hardy Fraction A (Fr.A; LIN- AA4+ Kit+ IL7Ra+ B220+), pro-B or Fr.B/C (IgM- CD19+ CD43+ CD24+), cycling pre-B or Fr.C' (IgM- CD19+ CD43+ CD24++), and resting pre-B or Fr.D (IgM- CD19+ CD43- CD24+) on the way to becoming immature B or Fr.E cells (IgM+ CD19+ CD43- CD24+; Figure 1A).9-11 Upon productive V(D)J rearrangement of the immunoglobulin (Ig) heavy gene locus, Fr.B/C progenitors, which express the pre-BCR components λ5 and VpreB1 (encoded by Igll1 and Vpreb1), form a functional pre-BCR.9,10 Pre-BCR signaling induces cell cycle entry and a burst of proliferation. Subsequently, cycling Fr.C' pre-B cells down-regulate the pre-BCR and exit the cycle to initiate Ig light chain rearrangement as resting Fr.D pre-B cells. This is a critical developmental transition, because B-cell progenitors have virtually unlimited proliferative potential. If unrestrained, B-cell progenitor proliferation can result in the accumulation of genetic lesions and malignant transformation toward B-cell–progenitor acute lymphoblastic leukemia.7-10 Ikzf1 expression steadily increases over the course of B-cell progenitor differentiation (www.immgen.org),12 and the Aiolos-encoding Ikaros paralog Ikzf3 is sharply up-regulated in response to pre-BCR signals13 (Figure 1B). Ikzf1 and Ikzf3 share a highly conserved set of N-terminal zinc fingers that confer DNA-binding specificity,14-16 regulate common target genes,17-19 cooperate to silence the expression of the pre-BCR genes Igll1 and Vpreb1, and facilitate the transition from pre-BCR–driven proliferation to cell cycle arrest, differentiation, and Ig light chain rearrangement by directly or indirectly regulating the expression of Myc, cell cycle genes, and Rag1 and Rag213,17,19-22 .
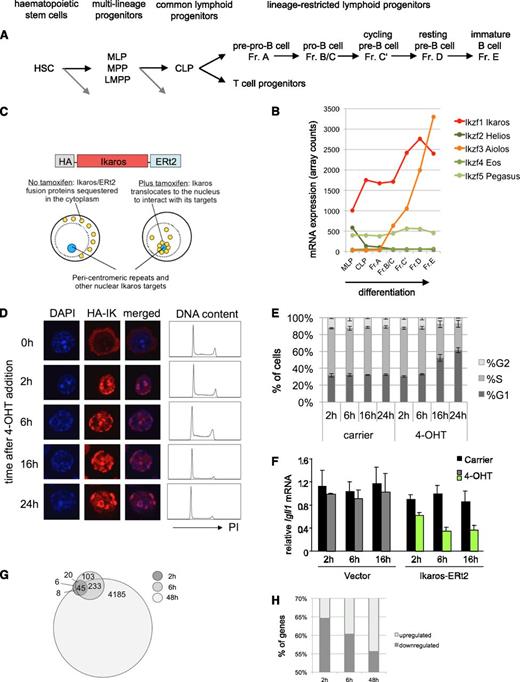
Ikaros-responsive genes in pre-B cells. (A) Scheme of adult B lymphocyte development in the context of hematopoiesis. Black arrows indicate developmental progression toward and within the lymphoid lineages, gray arrows indicate alternative lineage choices that are not depicted here. (B) Regulation of the Ikaros gene family during hematopoiesis and B-cell development. Data are normalized array counts from www.immgen.org. (C) Schematic representation of the inducible Ikaros construct and the experimental system. Ikaros-ERt2 is expressed at levels comparable with endogenous Ikaros (supplemental Figure 1D) and tethered in the cytoplasm until the addition of 4-hydroxy tamoxifen (4-OHT) triggers its release into the nucleus, where Ikaros is visible at DAPI-dense pericentromeric heterochromatin.30 (D) Nuclear translocation of inducible Ikaros (2 h) precedes G1 arrest (16 h). Immunofluorescence images of DAPI and hemagglutinin-Ikaros (HA-Ik) are representative of 2 experiments. DNA content was determined by flow cytometry of propidium iodide-stained cells at the indicated time points (representative of 3 experiments). (E) Bar graphs show the percentage of G1, S, and G2/M cells at the indicated time points (n = 3, mean ± SD). (F) Repression of the Ikaros target gene Igll1 is initiated within 2 h of Ikaros induction as determined by quantitative reverse transcription-polymerase chain reaction (n = 5, mean ± SD). (G) The relationship between Ikaros-responsive genes at 2 h, 6 h, and 48 h. (H) Genes that change their expression in response to Ikaros at early time points are more likely to be down-regulated.
Ikaros-responsive genes in pre-B cells. (A) Scheme of adult B lymphocyte development in the context of hematopoiesis. Black arrows indicate developmental progression toward and within the lymphoid lineages, gray arrows indicate alternative lineage choices that are not depicted here. (B) Regulation of the Ikaros gene family during hematopoiesis and B-cell development. Data are normalized array counts from www.immgen.org. (C) Schematic representation of the inducible Ikaros construct and the experimental system. Ikaros-ERt2 is expressed at levels comparable with endogenous Ikaros (supplemental Figure 1D) and tethered in the cytoplasm until the addition of 4-hydroxy tamoxifen (4-OHT) triggers its release into the nucleus, where Ikaros is visible at DAPI-dense pericentromeric heterochromatin.30 (D) Nuclear translocation of inducible Ikaros (2 h) precedes G1 arrest (16 h). Immunofluorescence images of DAPI and hemagglutinin-Ikaros (HA-Ik) are representative of 2 experiments. DNA content was determined by flow cytometry of propidium iodide-stained cells at the indicated time points (representative of 3 experiments). (E) Bar graphs show the percentage of G1, S, and G2/M cells at the indicated time points (n = 3, mean ± SD). (F) Repression of the Ikaros target gene Igll1 is initiated within 2 h of Ikaros induction as determined by quantitative reverse transcription-polymerase chain reaction (n = 5, mean ± SD). (G) The relationship between Ikaros-responsive genes at 2 h, 6 h, and 48 h. (H) Genes that change their expression in response to Ikaros at early time points are more likely to be down-regulated.
Ikaros chromatin immunoprecipitation and high throughput sequencing (ChIP-seq) data are available for human hematopoietic progenitors23 and mouse thymocytes,24,25 where Ikaros and Aiolos show highly similar binding.25 However, the genome-wide integration of Ikaros binding and gene expression has not been achieved in B-cell progenitors due to a number of confounding factors. First, in vitro site selection experiments have yielded only a simple consensus sequence, (G)GGAA(A), which resembles that of Ets proteins.26,27 The number of such motifs in the genome is high and limits the utility of in silico target predictions. Second, Ikzf1 deletion results in severe developmental disturbances,3,28 which have precluded the use of constitutive Ikaros knockouts for the identification of Ikaros targets in developing B cells, and conditional Ikzf1 alleles are not available at present. Third, Ikaros is an inhibitor of cell cycle progression,29 and previous analyses of Ikaros target genes in this pathway by retroviral gene transfer were only possible at time points when G1 arrest had already occurred.18
To overcome these limitations, we have combined Ikaros ChIP-seq with gene expression studies at enhanced temporal resolution. In addition to confirming several previously described Ikaros target genes, the global identification of Ikaros target genes allowed us to address questions that could not be approached by single gene studies. First, we examined biological pathways that are important for pre-B–cell differentiation and found that Ikaros target genes were highly enriched in pre-B–cell receptor signaling, cell cycle, and V(D)J recombination. Second, we determined the role of Ikaros target genes in B-cell lineage specification and developmental progression of B-cell progenitors along the B-cell lineage. To this end, we integrated our ChIP-seq and gene expression data with gene expression changes during hematopoietic development (www.immgen.org).12 Remarkably, Ikaros-regulated genes accounted for more than one-half of all genes that were up-regulated in vivo at the transition from CLP toward B-cell progenitors, demonstrating that Ikaros regulates B lineage-associated genes from the earliest stages of B-cell differentiation. Furthermore, increased Ikaros expression was sufficient to drive the majority of gene expression changes associated with the progression from the cycling to the resting pre-B–cell stage (Fr.C' to Fr.D) in vivo. These results provide an explanation for the essential role of Ikaros in B-cell lineage specification and demonstrate that the expression of Ikaros transcription factors, which is incremental during pre-B–cell development13 (Figure 1B), drives the differentiation from cycling to resting stages of pre-B–cell development.
Materials and methods
Cells and retroviral infections
The B3 pre-B–cell line30 was transduced27 with mouse stem cell virus retroviral vectors encoding wild-type Ikaros or Aiolos tagged with a hemagglutinin (HA) epitope followed by an internal ribosomal entry site (IRES) and GFP (HA-Ikaros-IRES-GFP, HA-Aiolos-IRES-GFP), the DNA binding-deficient Ikaros mutant 159A (HA-159A Ikaros-IRES-GFP), or control vector (IRES-GFP). B3 cells containing inducible Ikaros (HA-Ikaros-ERt2-IRES-GFP) were treated with 4-hydroxy-tamoxifen for the indicated times and ERt2-IRES-GFP served as the control vector. Animals were used under the authority of a UK Home Office license, PPL 70/6845.
Flow cytometry, immunofluorescence staining and cell cycle analysis
Cells were sorted based on GFP levels using FACSDiva or FACSAria instruments (Beckton Dickinson, Oxford, UK). Immunofluorescence staining with anti-HA-TRITC (Santa Cruz Biotechnology, Heidelberg, Germany) and 4,6 diamidino-2-phenylindole (DAPI) (Vector Laboratories, Peterborough, UK) was as described.13 DNA content was determined on a LSRII flow cytometer (Beckton Dickinson) after incubation in PBS, 50 ug/mL PI, 0.05% NP40, and 1 mg/mL RNaseA for 30 min on ice.
Please see supplemental Methods for a detailed description of gene expression profiling, ChIP, ChIP-seq, motif analysis, and meta-analysis of published gene expression data.
Results
A system for inducible Ikaros expression
To identify Ikaros-regulated genes in pre-B cells genome wide and to probe the significance of increasing Ikzf1 and Ikzf3 expression during B-cell differentiation12,13,17,18 (www.immgen.org; Figure 1B), we used retroviral gene transfer to express either wild-type Ikaros (HA-Ikaros-IRES-GFP) or the dominant negative Ikaros DNA-binding mutant 159A (HA-159A Ikaros-IRES-GFP)13 (supplemental Figure 1A). We chose the mouse pre-B–cell line B3 as our experimental system, because B3 cells resemble proliferating pre-B (Fr.C’) cells in their gene expression profile13,30 (supplemental Figure 1B). As expected for proliferating pre-B cells, B3 cells express moderate levels of endogenous Ikaros (supplemental Figure 1A), but little or no endogenous Aiolos (supplemental Figure 1C), and respond to experimentally enforced expression of Ikaros by cell cycle arrest (see below) and down-regulation of the known Ikaros target gene Igll1 (supplemental Figure 1A).13 Retrovirally transduced B3 cells were sorted for GFP expression and analyzed by Affymetrix gene expression arrays. We found that 4471 of 16 291 genes were differentially expressed between B3 cells transduced 48 h earlier with wild-type Ikaros vs 159A Ikaros and therefore appeared to be directly or indirectly regulated by Ikaros (1982 were up-regulated and 2486 were down-regulated; supplemental Excel File 1). To focus on more immediate Ikaros targets, we developed an inducible system that allows the control of Ikaros levels in the nucleus. Pre-formed Ikaros-ERt2 fusion proteins (Figure 1C) are expressed at levels comparable with endogenous Ikaros (supplemental Figure 1D). Ikaros-ERt2 fusion proteins were retained in the cytoplasm until the addition of 4-hydroxy-tamoxifen, which allows their release and nuclear translocation (Figure 1C-D). As previously reported,13,17,19,29 Ikaros led to an accumulation of cells in the G1 phase of the cell cycle, but in contrast to these earlier studies, the inducible system allowed us to measure Ikaros-induced changes in gene expression prior to cell cycle inhibition, which became noticeable 16 h after induction (Figure 1D-E). Down-regulation of the known Ikaros target gene Igll1 was observed within 2 h of Ikaros induction (Figure 1F). Using this system and Affymetrix gene expression arrays, we identified 79 and 401 Ikaros-regulated genes at 2 and 6 h after induction, respectively (Figure 1G; Supplementary Excel file 1), where the proportion of down-regulated transcripts was higher than at late time points (Figure 1H).
Genome-wide identification of Ikaros binding sites in pre-B cells
ChIP-seq with antibodies to the C terminus of endogenous Ikaros in B3 cells and with anti-HA in B3 cells transduced with epitope-tagged HA-Ikaros identified 11 349 high-confidence binding sites that were defined by the overlap between endogenous and HA-Ikaros peaks (Figure 2A). Ikaros ChIP-seq with antibodies to the C terminus of endogenous Ikaros in primary pre-B cells gave results that were similar to B3 cells, with 65.6% of B3 cell peaks also present in primary pre-B cells and 82% Ikaros-peaked genes in B3 cells also peaked in primary pre-B cells (Figure 2B).
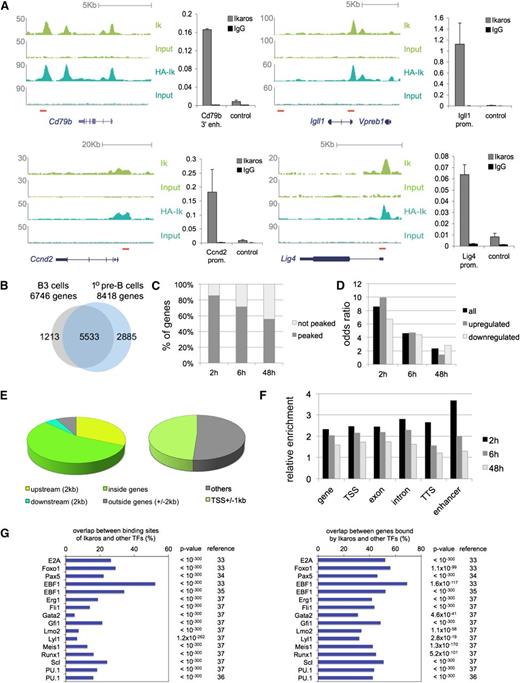
Genome-wide mapping of Ikaros binding sites in pre-B cells. (A) Ikaros ChIP-seq data for Cd79b, Igll1/Vbreb1, Ccnd2, and Lig4 with ChIP- polymerase chain reaction (PCR) validation. Endogenous Ikaros (top) and HA-Ikaros (bottom) tracks for B3 cells are shown alongside corresponding input samples. Red horizontal bars indicate the position of primers used for ChIP-PCR validation. Prom, promoter; enh, enhancer; control: Chr16: 16 853 772 near Igll1. (B) Genes bound by Ikaros in B3 cells (gray) and primary pre-B cells (blue) overlap significantly (P < 10−16, hypergeometric test, gene body plus 2 kb upstream of the transcription start site). (C) The percentage of Ikaros-regulated genes that is bound by Ikaros at 2, 6, and 48 h. (D) Odds ratios of Ikaros binding at differentially expressed genes after 2, 6, and 48 h. (E) Most Ikaros binding sites map in and around genes. (F) Enrichment of Ikaros binding at genomic features of Ikaros-responsive genes (gene = TSS to TTS ± 2 kb), TSS (±1 kb), TTS (+2 kb), and enhancers. Enhancers were defined by the presence in pro-B cells of the enhancer mark H3K4me1 or –me2 and the binding of Ikaros in addition to at least one of the transcription factors E2A, EBF1, or FOXO1 within a 1-kb window and the absence of the promoter mark H3K4me3 within 3 kb. These criteria were met by 771 putative enhancers surrounding 499 genes (3.06% of 16291 genes). (G) Experimental co-occurrence of Ikaros binding with other hematopoietic transcription factors (TFs).
Genome-wide mapping of Ikaros binding sites in pre-B cells. (A) Ikaros ChIP-seq data for Cd79b, Igll1/Vbreb1, Ccnd2, and Lig4 with ChIP- polymerase chain reaction (PCR) validation. Endogenous Ikaros (top) and HA-Ikaros (bottom) tracks for B3 cells are shown alongside corresponding input samples. Red horizontal bars indicate the position of primers used for ChIP-PCR validation. Prom, promoter; enh, enhancer; control: Chr16: 16 853 772 near Igll1. (B) Genes bound by Ikaros in B3 cells (gray) and primary pre-B cells (blue) overlap significantly (P < 10−16, hypergeometric test, gene body plus 2 kb upstream of the transcription start site). (C) The percentage of Ikaros-regulated genes that is bound by Ikaros at 2, 6, and 48 h. (D) Odds ratios of Ikaros binding at differentially expressed genes after 2, 6, and 48 h. (E) Most Ikaros binding sites map in and around genes. (F) Enrichment of Ikaros binding at genomic features of Ikaros-responsive genes (gene = TSS to TTS ± 2 kb), TSS (±1 kb), TTS (+2 kb), and enhancers. Enhancers were defined by the presence in pro-B cells of the enhancer mark H3K4me1 or –me2 and the binding of Ikaros in addition to at least one of the transcription factors E2A, EBF1, or FOXO1 within a 1-kb window and the absence of the promoter mark H3K4me3 within 3 kb. These criteria were met by 771 putative enhancers surrounding 499 genes (3.06% of 16291 genes). (G) Experimental co-occurrence of Ikaros binding with other hematopoietic transcription factors (TFs).
Of 6746 genes with at least one high-confidence Ikaros binding site, 5732 were represented on Affymetrix microarrays. The data in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession numbers GSE38110 (gene expression data for B3 cells), GSE42551 (gene expression data for primary pre-B cells), GSE38169 (ChIP-seq data for B3 cells), and GSE42462 (ChIP-seq data for primary pre-B cells).
Importantly, most genes that were regulated by Ikaros were also physical targets of Ikaros. The odds ratios for binding of both endogenous Ikaros and HA-Ikaros were 8.55, 4.58, and 2.33 for genes regulated at 2, 6, and 48 h, respectively (Table 1). Genes bound by HA-Ikaros only (ie, appeared to be selectively targeted by increased Ikaros levels) were not enriched for regulation by Ikaros (odds ratios 1.04, 1.07, and 1.41 for genes regulated at 2, 6 and 48 h, respectively), consistent with our previous analysis of the pre-BCR component Igll1, which was already bound by Ikaros in proliferating pre-B cells and then down-regulated by the increased expression of Ikaros factors in resting pre-B cells.13
Ikaros binding to differentially expressed genes in pre-B cells
| . | All . | Up-regulated . | Down-regulated . |
|---|---|---|---|
| Ikaros-responsive genes at 2 h | 79 | 28 | 51 |
| Ikaros binding observed | 65 (82.3%) | 22 (78.6%) | 43 (84.3%) |
| Ikaros binding expected | 28 | 10 | 18 |
| P value | 9.90 × 10−19 | 4.55 × 10−7 | 5.08 × 10−14 |
| Odds ratio | 8.55 | 6.75 | 9.90 |
| Ikaros-responsive genes at 6 h | 401 | 159 | 242 |
| Ikaros binding observed | 286 (71.3%) | 112 (70.4%) | 174 (71.9%) |
| Ikaros binding expected | 141 | 56 | 85 |
| P value | 5.88 × 10−51 | 2.35 × 10−20 | 2.39 × 10−32 |
| Odds ratio | 4.58 | 4.39 | 4.71 |
| Ikaros-responsive genes at 48 h | 4471 | 1982 | 2486 |
| Ikaros binding observed | 2497 (55.8%) | 1195 (60.3%) | 1302 (41.5%) |
| Ikaros binding expected | 1573 | 697 | 875 |
| P value | 2.16 × 10−246 | 1.43 × 10−131 | 7.52 × 10−82 |
| Odds ratio | 2.33 | 2.80 | 1.398 |
| . | All . | Up-regulated . | Down-regulated . |
|---|---|---|---|
| Ikaros-responsive genes at 2 h | 79 | 28 | 51 |
| Ikaros binding observed | 65 (82.3%) | 22 (78.6%) | 43 (84.3%) |
| Ikaros binding expected | 28 | 10 | 18 |
| P value | 9.90 × 10−19 | 4.55 × 10−7 | 5.08 × 10−14 |
| Odds ratio | 8.55 | 6.75 | 9.90 |
| Ikaros-responsive genes at 6 h | 401 | 159 | 242 |
| Ikaros binding observed | 286 (71.3%) | 112 (70.4%) | 174 (71.9%) |
| Ikaros binding expected | 141 | 56 | 85 |
| P value | 5.88 × 10−51 | 2.35 × 10−20 | 2.39 × 10−32 |
| Odds ratio | 4.58 | 4.39 | 4.71 |
| Ikaros-responsive genes at 48 h | 4471 | 1982 | 2486 |
| Ikaros binding observed | 2497 (55.8%) | 1195 (60.3%) | 1302 (41.5%) |
| Ikaros binding expected | 1573 | 697 | 875 |
| P value | 2.16 × 10−246 | 1.43 × 10−131 | 7.52 × 10−82 |
| Odds ratio | 2.33 | 2.80 | 1.398 |
More than 80% of genes that were differentially expressed 2 h after Ikaros induction were bound by Ikaros (Table 1; Figure 2C). With 84.3%, enrichment for Ikaros binding was particularly high for genes that were repressed by Ikaros (odds ratio = 9.9, P value = 6.08 × 10−14; Table 1; Figure 2D). The frequency of Ikaros-regulated genes with Ikaros binding was 70% at 6 h and 56% at 48 h (Table 1; Figure 2C). Hence, genes responsive to Ikaros at early time points were more likely physical targets of Ikaros binding, even though the enrichment at 48 h was still highly significant (P = 2.16 × 10−246). Consistent with a role for Ikaros in gene activation and repression, both up- and down-regulated gene sets were strongly enriched for Ikaros binding (Figure 2D).
Genomic features of gene regulation by Ikaros
Most Ikaros binding sites mapped within 2 kb of annotated genes (89.9%, Figure 2E) and nearly one-half (48.9%) were within 1 kb of transcription start sites (>23-fold enrichment over random expectation; Figure 2E), indicating that Ikaros binding is promoter-centric in B-cell progenitors. Promoter binding correlated with differential expression at 2, 6, and 48 h (P = 4.14 × 10−13, 7.32 × 10−38, and 2.93 × 10−237, respectively) to a similar degree as Ikaros binding elsewhere in genes and at putative enhancers (Figure 2F).
DNA sequence motifs and colocalization with other transcription factors
Most in vivo Ikaros binding motifs were variations of GGAA (supplemental Figure 2A motifs 1-5 and 10) as previously inferred by in vitro binding site selection studies.26,31 Four-basepair motifs occur on average every 256 nucleotides, or ∼107 times in the nonrepeat fraction of the mouse genome. Our estimate of ∼104 Ikaros binding sites suggests that ∼1 in 103 GGAA motifs are selectively occupied by Ikaros in vivo. Analysis of sequences adjacent to GGAA motifs within Ikaros ChIP-seq peaks identified E2A, Runx, and Lmo2 consensus sites, as well as sites for paired box (Pax) transcription factors, nuclear receptors (VDR, GR), AP-1, and the macrophage-associated factor Maf (supplemental Figure 2B).
Exceptions to the GGAA theme included a CAGCTG sequence (supplemental Figure 2A motif 9), which resembles an E2A motif identified in B-cell progenitors, 33 and a palindrome of GGGA with appropriate spacing to form an EBF1 consensus motif (supplemental Figure 2A motif 7). Both E2A and EBF1 have essential functions in B-cell development,9,10 and the identification of motifs recognized by these factors at sites of Ikaros binding suggested the possibility of colocalization between Ikaros and other transcription factors. We therefore compared our experimentally defined Ikaros binding sites with published EBF1, E2A, and Pax5 binding data.33,34 Remarkably, 52% of EBF1 sites and 26% of E2A sites in pro-B cells and 22% of Pax5 sites in pre-B cells34 overlapped with Ikaros peaks in B3 cells, as did 34.1% of EBF1 sites from an independent study35 (Figure 2G left; supplemental Table 1). With 29%, highly significant overlap was seen between Ikaros and Foxo1, another important factor for B-cell development (Figure 2G left; supplemental Table 1).33,35 Gene-centric analysis showed that 52% of E2A target genes, 69% of EBF1 target genes, 46% of Pax5 target genes, and 56% of Foxo1 target genes were also bound by Ikaros (Figure 2G right; supplemental Table 2). Moreover, binding of PU.1 in mature B cells36 and in murine HPC7 progenitor cells37 was highly correlated with Ikaros binding in B3 cells, as was binding of the transcription factors Erg1, Fli1, Gata2, Gfi1b, Lmo2, Lyl1, Meis1, Runx1, and Tal1/Scl (Figure 2G; supplemental Tables 1-2). This was the case even though Tal1/Scl, Meis1, and Lmo2 are preferentially expressed at early stages of hematopoiesis, rather than in pre-B cells from which our Ikaros binding maps were derived. PU.1, Erg1, and Fli1 are Ets factors that share the GGAA core recognition sequence with Ikaros proteins, but because only a small fraction of GGAA motifs are occupied by Ikaros in vivo (see above), sequence specificity alone does not explain the observed colocalization frequencies. We conclude that Ikaros occupies many of the same sites and targets many of the same genes as other hematopoietic transcription factors, including E2A, EBF1, and Foxo1.
Enrichment of Ikaros target genes in (pre)-B–cell receptor signaling, cell cycle progression, as well as V(D)J recombination
Ikaros regulated the expression of transcription factor genes important for the biology of progenitors (Lmo2, Lyl1) and B cells (Foxo1, Foxp1, Irf4, Irf8, Gfi1b, Tcf3; supplemental Excel File 2). In contrast to earlier work in multipotent progenitors,38 we saw no regulation of Sfpi1, no binding and no regulation of Cebpa, and no binding of Gfi1, suggesting that these genes may not be direct Ikaros targets in pre-B cells. Ikaros did, however, bind to and repress Gfi1b (see below). In addition to sequence-specific transcription factors, Ikaros regulated components of the general transcription machinery (such as mediator and RNA polymerase subunits Polr1a, -b, -c, -d, Polr2d, -e, -f, -g, -h, -j, -l, Polr3d, -e, and -h), translation (including ribosome biogenesis and translation factors), and metabolism (supplemental Excel File 2).
In contrast to single-gene studies, the global identification of Ikaros targets allowed the interrogation of entire biological pathways that are critical for B-cell progenitor differentiation. We found that Ikaros-regulated genes were highly represented in (pre)-B–cell receptor signaling, cell cycle progression, as well as V(D)J recombination (Figures 3-5).
Ikaros-induced gene expression changes relating to pre-BCR signaling. (A) Schematic representation of genes in the pre-BCR signaling pathway. Genes are in italics and the corresponding proteins are listed where they are better known than the genes. The cell membrane, nuclear membrane, and the pre-BCR are outlined. Orange, gold, and yellow mark significant mRNA expression changes at the indicated times and an asterisk shows that expression changes have been verified by quantitative reverse transcription-polymerase chain reaction (RT-PCR). Genes with a black outline are bound by Ikaros in ChIP experiments (±2 kb, see below). A gray background marks pathway components that did not change expression. (B) RT-PCR validation of Ikaros-induced gene expression changes related to pre-BCR signaling in B3 cells (mean ± SD; n = 3 independent biological replicates; * P < .05, ** P < .01, *** P < .001; Student t test). (C) RT-PCR validation of Aiolos-induced gene expression changes related to pre-BCR signaling in B3 cells (mean ± SD; n = 3 independent biological replicates; * P < .05, ** P < .01, *** P < .001; Student t test). (D) RT-PCR validation of Ikaros-induced gene expression changes related to pre-BCR signaling in primary pre-B cells (mean ± SD; n = 3 independent biological replicates; * P < .05, ** P < .01, *** P < .001; Student t test). The quantitative differences between Ikaros effects in B3 cells vs primary pre-B cells are due to the lower level of Ikaros expression achieved in primary pre-B cells (not shown). (E) Ikaros controls the expression of Syk, a key kinase upstream of SLP65 (encoded by Blnk) and thereby directs pre-BCR (or BCR-ABL) signals toward SLP65.
Ikaros-induced gene expression changes relating to pre-BCR signaling. (A) Schematic representation of genes in the pre-BCR signaling pathway. Genes are in italics and the corresponding proteins are listed where they are better known than the genes. The cell membrane, nuclear membrane, and the pre-BCR are outlined. Orange, gold, and yellow mark significant mRNA expression changes at the indicated times and an asterisk shows that expression changes have been verified by quantitative reverse transcription-polymerase chain reaction (RT-PCR). Genes with a black outline are bound by Ikaros in ChIP experiments (±2 kb, see below). A gray background marks pathway components that did not change expression. (B) RT-PCR validation of Ikaros-induced gene expression changes related to pre-BCR signaling in B3 cells (mean ± SD; n = 3 independent biological replicates; * P < .05, ** P < .01, *** P < .001; Student t test). (C) RT-PCR validation of Aiolos-induced gene expression changes related to pre-BCR signaling in B3 cells (mean ± SD; n = 3 independent biological replicates; * P < .05, ** P < .01, *** P < .001; Student t test). (D) RT-PCR validation of Ikaros-induced gene expression changes related to pre-BCR signaling in primary pre-B cells (mean ± SD; n = 3 independent biological replicates; * P < .05, ** P < .01, *** P < .001; Student t test). The quantitative differences between Ikaros effects in B3 cells vs primary pre-B cells are due to the lower level of Ikaros expression achieved in primary pre-B cells (not shown). (E) Ikaros controls the expression of Syk, a key kinase upstream of SLP65 (encoded by Blnk) and thereby directs pre-BCR (or BCR-ABL) signals toward SLP65.
Ikaros-induced gene expression changes relating to the cell cycle. (A) Symbols as in Figure 3. (B) RT-PCR validation of Ikaros-induced gene expression changes related to the cell cycle in B3 cells (mean ± SD; n = 3 independent biological replicates; * P < .05, ** P < .01; Student t test). (C) RT-PCR validation of Aiolos-induced gene expression changes related to the cell cycle in B3 cells (mean ± SD; n = 3 independent biological replicates; *** P < .001; Student t test). (D) RT-PCR validation of Ikaros-induced gene expression changes related to the cell cycle in primary pre-B cells (mean ± SD; n = 3 independent biological replicates; *** P < .001; Student t test).
Ikaros-induced gene expression changes relating to the cell cycle. (A) Symbols as in Figure 3. (B) RT-PCR validation of Ikaros-induced gene expression changes related to the cell cycle in B3 cells (mean ± SD; n = 3 independent biological replicates; * P < .05, ** P < .01; Student t test). (C) RT-PCR validation of Aiolos-induced gene expression changes related to the cell cycle in B3 cells (mean ± SD; n = 3 independent biological replicates; *** P < .001; Student t test). (D) RT-PCR validation of Ikaros-induced gene expression changes related to the cell cycle in primary pre-B cells (mean ± SD; n = 3 independent biological replicates; *** P < .001; Student t test).
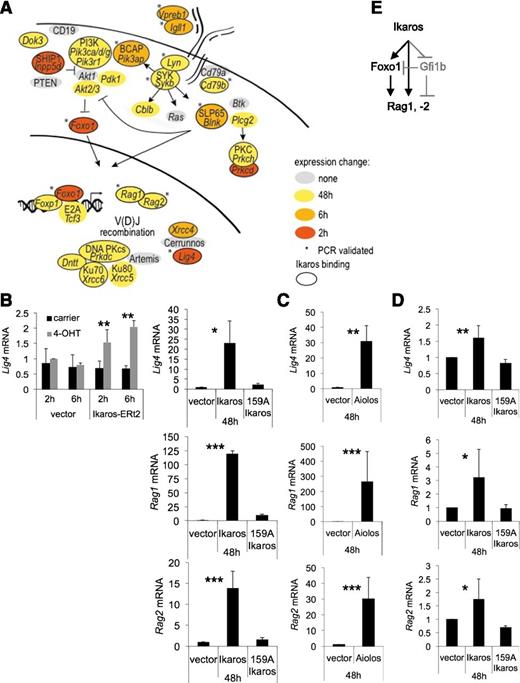
Ikaros-induced gene expression changes relating to V(D)J recombination. (A) Symbols as in Figure 3. (B) RT-PCR validation of Ikaros-induced gene expression changes related to V(D)J recombination in B3 cells (mean ± SD; n = 3 independent biological replicates; * P < .05, ** P < .01, *** P < .001; Student t test). (C) RT-PCR validation of Aiolos-induced gene expression changes related to V(D)J recombination in B3 cells (mean ± SD; n = 3 independent biological replicates; ** P < .01, *** P < .001; Student t test). (D) RT-PCR validation of Ikaros-induced gene expression changes related to V(D)J recombination in primary pre-B cells (mean ± SD; n = 3 independent biological replicates; * P < .05, ** P < .01; Student t test). (E) Model for the regulation of Rag1 and -2 through an Ikaros-based feed-forward loop involving Foxo1. Note that the design of feed-forward loops introduces a time delay so that not all targets of an upstream transcription factor will respond immediately to its expression. The model includes the Ikaros-regulated gene Gfi1b, which negatively regulates Rag expression both directly and indirectly through repression of Foxo1.
Ikaros-induced gene expression changes relating to V(D)J recombination. (A) Symbols as in Figure 3. (B) RT-PCR validation of Ikaros-induced gene expression changes related to V(D)J recombination in B3 cells (mean ± SD; n = 3 independent biological replicates; * P < .05, ** P < .01, *** P < .001; Student t test). (C) RT-PCR validation of Aiolos-induced gene expression changes related to V(D)J recombination in B3 cells (mean ± SD; n = 3 independent biological replicates; ** P < .01, *** P < .001; Student t test). (D) RT-PCR validation of Ikaros-induced gene expression changes related to V(D)J recombination in primary pre-B cells (mean ± SD; n = 3 independent biological replicates; * P < .05, ** P < .01; Student t test). (E) Model for the regulation of Rag1 and -2 through an Ikaros-based feed-forward loop involving Foxo1. Note that the design of feed-forward loops introduces a time delay so that not all targets of an upstream transcription factor will respond immediately to its expression. The model includes the Ikaros-regulated gene Gfi1b, which negatively regulates Rag expression both directly and indirectly through repression of Foxo1.
The pre-BCR consists of the Ig heavy chain and the surrogate light chain components λ5 and VpreB (encoded by Igll1 and Vpreb1) and is coupled to intracellular signaling pathways by a heterodimer of Igα and Igβ (encoded by Cd79a and Cd79b) via the tyrosine kinases Syk (encoded by Sykb) and Lyn. In addition to the pre-BCR components Igll1 and Vpreb1, we identified the signal transducer Cd79b and the downstream signaling components Lyn, Sykb, Blnk, Inpp5d (which encodes SHIP1), Cblb, the PI3K pathway components Pik3ap (encoding Bcap), Pik3ca, Pik3cd, Pik3cg, and Pik3r1, as well as the kinases Akt2 and Pdk1 as Ikaros-regulated genes (Figure 3A; supplemental Excel File 1). Quantitative reverse transcription-polymerase chain reaction (RT-PCR) confirmed the regulation of key Ikaros targets in the (pre)-B–cell receptor signaling pathway by Ikzf1 (Figure 3B) and Ikzf3 (Figure 3C; supplemental Figure 1C) and in primary pre-B cells (Figure 3D). Of particular interest was the upregulation of Blnk, which encodes SLP65, a protein essential for the termination of pre-BCR signaling, and its upstream regulator Syk (encoded by Sykb) by Ikaros and Aiolos (Figure 3E). SLP-65 is a key substrate of Syk and cooperates with BTK and PLCγ2 to regulate Irf4, Irf8, and Ikzf3. Irf4 and Foxo1 were among the transcription factor genes that are bound and regulated by Ikaros (Figure 3A; supplemental Excel File 1).
The termination of pre-BCR signaling releases FOXO proteins from Akt/PKB-mediated repression, stabilizes p27, and facilitates the transition from proliferating to resting pre-B cells. This involves the regulation of transcripts, including Foxo1, Ccnd2 (cyclin D2), Ccnd3 (cyclin D3), Cdk6, CDK inhibitors of the INK4 family (Cdkn2a, -b, -c, and -d), and the KIP family members Cdkn1a (p21) and Cdkn1b (p27). Ikaros down-regulated cyclins Ccnd2 and Ccnd3 and the cyclin-dependent kinase Cdk6 and up-regulated the cell cycle inhibitors Cdkn2a, Cdkn1a, and Cdkn1b (Figure 4A). Among the downstream cell cycle effectors regulated by Ikaros were E2F (E2f1, -2, -3, -5, -7, and -8), Tfdp1 (DP-1), and the chromatin modifiers Hdac8, Suv39h1, and Suv39h2. Ikaros bound and regulated Myc,17,18 Foxo1, and Smad3 (Figure 4A; supplemental Excel File 1). Quantitative RT-PCR confirmed the impact of Ikaros (Figure 4B) and Aiolos (Figure 4C) in B3 cells and primary pre-B cells (Figure 4D).
Importantly, Ccnd2, Cdk6, Cdkn1a, Cdkn1b, Myc, Foxo1, and Smad3 were significantly regulated 2 or 6 h after Ikaros induction, that is prior to Ikaros-mediated cell cycle arrest (Figure 1D-E), and therefore represent candidate genes for mediating Ikaros-induced cell cycle arrest.
Developmentally regulated cell cycle arrest limits the expansion of B-cell progenitors and facilitates Ig light chain rearrangement and B-cell differentiation. Ikaros-regulated transcripts in this pathway included the recombinases Rag1 and -2, the nonhomologous end joining components Xrcc4, Xrcc6, and Lig4, and the upstream regulators Foxo1, Foxp1, and Tcf3 (encoding E2A; Figure 5A). Foxo1, Foxp1, E2A, and Ikaros target the Erag recombination-activating gene enhancer10,32 to increase transcription of Rag1 and Rag2, and RAG proteins are stabilized by G1 arrest. IRF4 and E2A promote Ig light chain locus transcription and accessibility.17 Quantitative RT-PCR confirmed the impact of Ikaros (Figure 5B) and Aiolos (Figure 5C) in B3 cells and primary pre-B cells (Figure 5D). Gfi1b represses Rag expression directly and indirectly (through repressing Foxo1).39 Our data that Ikaros induces Foxo1 and Foxp1 and represses Gfi1b delineate a feed-forward loop that regulates Rag1 and Rag2 (Figure 5E). Feed-forward loops slow down responses,40 explaining why not all direct targets of Ikaros show regulation at early time points.
Ikaros target genes are differentially expressed in the context of hematopoiesis, lymphocyte lineage choice, and B-cell differentiation. (A) Ikaros target genes are differentially expressed during B-cell lineage specification in vivo. Vertical arrows describe gene expression changes at the respective developmental transitions (up for up-regulation, down for down-regulation). Left: arrow color indicates if Ikaros-regulated genes are enriched among up- and down-regulated genes in vivo (see color scale). Right: arrow color indicates if Ikaros-bound genes are enriched among up- and down-regulated genes in vivo. Supplemental Excel File 4 lists the genes that are regulated by Ikaros in B3 cells and differentially expressed during B-cell lineage specification. (B) Ikaros target genes are differentially expressed during the transition from proliferating to resting pre-B cells in vivo. Arrows describe gene expression changes at the developmental transition from the cycling to resting pre-B cells (up for up-regulation, down for down-regulation). Left: arrow color indicates if Ikaros-regulated genes are enriched among up- and down-regulated genes in vivo (see color scale). Right: arrow color indicates if Ikaros-bound genes are enriched among up- and down-regulated genes in vivo. Supplemental Excel File 4 lists the genes that are regulated by Ikaros in B3 cells and differentially expressed during the progression from Fr.C' to Fr.D. (C) Ikaros expression in B3 cells captures most gene expression changes that occur during the transition from proliferating to resting pre-B cells in vivo. The Venn diagram on the right shows down-regulated genes. (D) Heat map of mean-centered T-statistics for Pearson correlations between B3 cells and successive stages of B-cell progenitor differentiation. Control B3 cells most resemble Fr. C’ proliferating pre-B cells and Ikaros-transduced B3 cells most resemble Fr. D resting pre-B cells. (E) Heat map of developmental stage-specific gene expression changes during B-cell differentiation in vivo (relative to the population average) and gene expression changes induced by Ikaros expression in B3 cells. A hierarchical tree based on Spearman's rank correlation shows pairwise distances between populations. (F) Primary pre-B cells expanded on ST2 feeder cells in the presence of interleukin-7 were transduced with Ikaros-IRES-GFP or IRES-GFP control vector as described13 and sorted for GFP expression 48 h later. Gene expression changes were determined using Affymetrix arrays for 2 biological replicates and results were overlaid with developmental gene expression data as described in A and B. Supplemental Excel File 5 lists the genes that are regulated by Ikaros in primary pre-B cells and differentially expressed during B-cell lineage specification and the progression from Fr.C' to Fr.D.
Ikaros target genes are differentially expressed in the context of hematopoiesis, lymphocyte lineage choice, and B-cell differentiation. (A) Ikaros target genes are differentially expressed during B-cell lineage specification in vivo. Vertical arrows describe gene expression changes at the respective developmental transitions (up for up-regulation, down for down-regulation). Left: arrow color indicates if Ikaros-regulated genes are enriched among up- and down-regulated genes in vivo (see color scale). Right: arrow color indicates if Ikaros-bound genes are enriched among up- and down-regulated genes in vivo. Supplemental Excel File 4 lists the genes that are regulated by Ikaros in B3 cells and differentially expressed during B-cell lineage specification. (B) Ikaros target genes are differentially expressed during the transition from proliferating to resting pre-B cells in vivo. Arrows describe gene expression changes at the developmental transition from the cycling to resting pre-B cells (up for up-regulation, down for down-regulation). Left: arrow color indicates if Ikaros-regulated genes are enriched among up- and down-regulated genes in vivo (see color scale). Right: arrow color indicates if Ikaros-bound genes are enriched among up- and down-regulated genes in vivo. Supplemental Excel File 4 lists the genes that are regulated by Ikaros in B3 cells and differentially expressed during the progression from Fr.C' to Fr.D. (C) Ikaros expression in B3 cells captures most gene expression changes that occur during the transition from proliferating to resting pre-B cells in vivo. The Venn diagram on the right shows down-regulated genes. (D) Heat map of mean-centered T-statistics for Pearson correlations between B3 cells and successive stages of B-cell progenitor differentiation. Control B3 cells most resemble Fr. C’ proliferating pre-B cells and Ikaros-transduced B3 cells most resemble Fr. D resting pre-B cells. (E) Heat map of developmental stage-specific gene expression changes during B-cell differentiation in vivo (relative to the population average) and gene expression changes induced by Ikaros expression in B3 cells. A hierarchical tree based on Spearman's rank correlation shows pairwise distances between populations. (F) Primary pre-B cells expanded on ST2 feeder cells in the presence of interleukin-7 were transduced with Ikaros-IRES-GFP or IRES-GFP control vector as described13 and sorted for GFP expression 48 h later. Gene expression changes were determined using Affymetrix arrays for 2 biological replicates and results were overlaid with developmental gene expression data as described in A and B. Supplemental Excel File 5 lists the genes that are regulated by Ikaros in primary pre-B cells and differentially expressed during B-cell lineage specification and the progression from Fr.C' to Fr.D.
Ikaros-regulated genes in the context of hematopoiesis and B-cell development
As an approach to evaluate the significance of Ikaros-mediated gene regulation during B-cell development, we next determined the proportion of Ikaros targets among genes that are differentially expressed at sequential stages of B-cell lineage specification and differentiation (www.immgen.org).
Ikaros-regulated genes were enriched among genes differentially expressed during the in vivo transition from MLP to CLP (Table 2; Figure 6A; supplemental Excel File 4). This was the case particularly for up-regulated genes (Table 2; Figure 6A; supplemental Excel File 4) and in the 2-h and 6-h data sets (Table 2; see supplemental Table 3 for a comparison with the gene expression clusters derived from hematopoietic stem and progenitor populations28 ). An even stronger enrichment for Ikaros targets was found among genes differentially expressed during the specification of the B-cell lineage (CLP to Fr.A; Table 2; Figure 6A; supplementary Excel File 4). Remarkably, this enrichment was entirely due to genes that were up-regulated at the CLP to Fr.A transition; more than one-half (53.7%) of all genes up-regulated at the CLP to Fr.A transition in vivo were regulated by Ikaros in pre-B cells (Table 2; Figure 6A; supplemental Excel File 4). Enrichment was consistent throughout the time course and was confirmed for Ikaros-bound as well as for Ikaros-regulated genes (Table 2). In stark contrast, Ikaros targets were not enriched and indeed were depleted among genes that were down-regulated at the CLP to Fr.A transition (Table 2; Figure 6A; supplemental Excel File 4). Interestingly, the opposite pattern emerged when we analyzed gene expression changes in cells that chose an alternative fate and progressed from CLPs toward the T-cell lineage: pre-B cell-defined Ikaros targets were underrepresented among genes up-regulated in DP thymocytes relative to CLPs (odds ratios were 0.59, 0.59, and 0.46 for the 2-, 6-, and 48-h Ikaros target sets, respectively; Table 2; Figure 6A; supplemental Excel File 4). Instead, Ikaros targets were strongly enriched for genes that were down-regulated in thymocytes relative to CLPs (Table 2; Figure 6A). Genes bound by Ikaros in pre-B cells were preferentially down-regulated during thymocyte differentiation (Table 2; Figure 6A; supplemental Excel File 4). We conclude that Ikaros targets defined in pre-B cells are highly overrepresented among genes that are differentially regulated at key steps of lymphoid progenitor differentiation and lymphoid lineage specification in vivo.
Differential expression and overlap with Ikaros-regulated genes
| . | Differential expression MLP to CLP . | Differential expression CLP to DP thymocytes . | ||||
|---|---|---|---|---|---|---|
| . | All . | Up-regulated . | Down-regulated . | All . | Up-regulated . | Down-regulated . |
| All genes (16 291) | 868 | 362 | 506 | 7753 | 3849 | 3904 |
| Overlap with Ikaros-regulated genes | ||||||
| 2-h Gene set (79) | 18 | 13 | 5 | 54 | 11 | 43 |
| P value | 2.23 x10−8 | 1.88 × 10−9 | 0.036 | 5.9 × 10−5 | 0.98 | 1.33 × 10−9 |
| Odds ratio | 4.346 | 7.644 | 2.048 | 1.44 | 0.59 | 2.29 |
| 6-h Gene set (401) | 64 | 34 | 30 | 256 | 56 | 200 |
| P value | 5.88 × 10−16 | 4.82 × 10−12 | 3.13 × 10−6 | 1.26 × 10−11 | 1.00 | 1.39 × 10−30 |
| Odds ratio | 3.154 | 4.108 | 2.497 | 1.35 | 0.59 | 2.14 |
| 48-h Gene set (4471) | 322 | 147 | 175 | 2464 | 567 | 1897 |
| P value | 7.55 × 1011 | 1.66 × 10−8 | 1.40 × 10−4 | 1.26 × 10−32 | 1 | 8.13 × 10−237 |
| Odds ratio | 1.559 | 1.808 | 1.398 | 1.23 | 0.46 | 2.50 |
| Overlap with Ikaros-bound genes | ||||||
| Bound gene set (5732) | 390 | 192 | 198 | 2842 | 899 | 1943 |
| P value | 4.71 × 10−10 | 7.70 × 10−13 | 0.027 | 8.36 × 10−5 | 1 | 1.21 × 10−103 |
| Odds ratio | 1.50 | 2.08 | 1.18 | 1.07 | 0.56 | 1.83 |
| All genes (16 291) | 2323 | 1095 | 1228 | 2496 | 831 | 1665 |
| Overlap with Ikaros-regulated genes | ||||||
| 2-h Gene set (79) | 21 | 16 | 5 | 23 | 12 | 11 |
| P value | 0.001 | 1.55 × 10−5 | 0.55 | 5.33 × 10−4 | 1.67 × 10−4 | 0.11 |
| Odds ratio | 1.872 | 3.043 | 0.839 | 1.91 | 3.01 | 1.36 |
| 6-h Gene set (401) | 117 | 84 | 33 | 169 | 46 | 123 |
| P value | 1.74 × 10−15 | 6.69 × 10−22 | 0.26 | 9.60 × 10−40 | 8.78 × 10−8 | 4.35 × 10−31 |
| Odds ratio | 2.101 | 3.292 | 1.094 | 2.879 | 2.322 | 3.161 |
| 48-h Gene set (4471) | 926 | 588 | 338 | 1523 | 397 | 1126 |
| P value | 2.93 × 10−45 | 3.22 × 10−81 | 0.46 | 0.00 | 4.26 × 10−38 | 6.75 × 10−290 |
| Odds ratio | 1.752 | 3.066 | 1.00 | 4.13 | 2.42 | 5.52 |
| Overlap with Ikaros-bound genes | ||||||
| Bound gene set (5732) | 1125 | 662 | 463 | 1283 | 472 | 811 |
| P value | 4.13 × 1046 | 3.59 × 10−70 | 0.026 | 1.52 × 10−73 | 2.24 × 10−39 | 1.96 × 10−33 |
| Odds ratio | 1.73 | 2.82 | 1.11 | 1.95 | 2.42 | 1.75 |
| . | Differential expression MLP to CLP . | Differential expression CLP to DP thymocytes . | ||||
|---|---|---|---|---|---|---|
| . | All . | Up-regulated . | Down-regulated . | All . | Up-regulated . | Down-regulated . |
| All genes (16 291) | 868 | 362 | 506 | 7753 | 3849 | 3904 |
| Overlap with Ikaros-regulated genes | ||||||
| 2-h Gene set (79) | 18 | 13 | 5 | 54 | 11 | 43 |
| P value | 2.23 x10−8 | 1.88 × 10−9 | 0.036 | 5.9 × 10−5 | 0.98 | 1.33 × 10−9 |
| Odds ratio | 4.346 | 7.644 | 2.048 | 1.44 | 0.59 | 2.29 |
| 6-h Gene set (401) | 64 | 34 | 30 | 256 | 56 | 200 |
| P value | 5.88 × 10−16 | 4.82 × 10−12 | 3.13 × 10−6 | 1.26 × 10−11 | 1.00 | 1.39 × 10−30 |
| Odds ratio | 3.154 | 4.108 | 2.497 | 1.35 | 0.59 | 2.14 |
| 48-h Gene set (4471) | 322 | 147 | 175 | 2464 | 567 | 1897 |
| P value | 7.55 × 1011 | 1.66 × 10−8 | 1.40 × 10−4 | 1.26 × 10−32 | 1 | 8.13 × 10−237 |
| Odds ratio | 1.559 | 1.808 | 1.398 | 1.23 | 0.46 | 2.50 |
| Overlap with Ikaros-bound genes | ||||||
| Bound gene set (5732) | 390 | 192 | 198 | 2842 | 899 | 1943 |
| P value | 4.71 × 10−10 | 7.70 × 10−13 | 0.027 | 8.36 × 10−5 | 1 | 1.21 × 10−103 |
| Odds ratio | 1.50 | 2.08 | 1.18 | 1.07 | 0.56 | 1.83 |
| All genes (16 291) | 2323 | 1095 | 1228 | 2496 | 831 | 1665 |
| Overlap with Ikaros-regulated genes | ||||||
| 2-h Gene set (79) | 21 | 16 | 5 | 23 | 12 | 11 |
| P value | 0.001 | 1.55 × 10−5 | 0.55 | 5.33 × 10−4 | 1.67 × 10−4 | 0.11 |
| Odds ratio | 1.872 | 3.043 | 0.839 | 1.91 | 3.01 | 1.36 |
| 6-h Gene set (401) | 117 | 84 | 33 | 169 | 46 | 123 |
| P value | 1.74 × 10−15 | 6.69 × 10−22 | 0.26 | 9.60 × 10−40 | 8.78 × 10−8 | 4.35 × 10−31 |
| Odds ratio | 2.101 | 3.292 | 1.094 | 2.879 | 2.322 | 3.161 |
| 48-h Gene set (4471) | 926 | 588 | 338 | 1523 | 397 | 1126 |
| P value | 2.93 × 10−45 | 3.22 × 10−81 | 0.46 | 0.00 | 4.26 × 10−38 | 6.75 × 10−290 |
| Odds ratio | 1.752 | 3.066 | 1.00 | 4.13 | 2.42 | 5.52 |
| Overlap with Ikaros-bound genes | ||||||
| Bound gene set (5732) | 1125 | 662 | 463 | 1283 | 472 | 811 |
| P value | 4.13 × 1046 | 3.59 × 10−70 | 0.026 | 1.52 × 10−73 | 2.24 × 10−39 | 1.96 × 10−33 |
| Odds ratio | 1.73 | 2.82 | 1.11 | 1.95 | 2.42 | 1.75 |
A key role for Ikaros proteins at the transition from the cycling to the resting pre-B cells stage
Next, we focused on the transition from the cycling to the resting stage of pre-B–cell development. Here, the Ikaros family member Aiolos is sharply up-regulated in response to pre-BCR signals relayed through SLP-6513 and cooperates with Ikaros in the silencing of pre-BCR components and Myc expression.13,18 We first examined the overlap between pre-B cell-defined Ikaros targets and genes that are differentially expressed at the transition from cycling to resting pre-B (Fr.C' to Fr.D) cells. We found a highly significant enrichment of pre-B cell-defined Ikaros targets at all time points (Table 2; Figure 6B; supplemental Excel File 4). A total of 61% of all genes that are differentially expressed genes at the Fr.C' to Fr.D transition in vivo were regulated by Ikaros in B3 cells (48 h; Table 2, Figure 6C; supplemental Excel File 4). The most striking enrichment was found for genes that were down-regulated during the transition from Fr.C' to Fr.D in vivo, more than two-thirds of which (67.6%) were regulated by Ikaros in B3 cells (Table 2; Figure 6C; supplemental Excel File 4). Of 1523 genes that were differentially expressed at the Fr.C' to Fr.D transition, 1415 changed in the same direction in vivo and in vitro (346 were up-regulated and 1069 down-regulated), which represents 92.9% concordance between in vivo and in vitro data sets. Enforced expression of Ikaros changes the global gene expression profile of B3 cells from one that resembled Fr.C' to one that more closely resembled Fr.D pre-B cells (Figure 6D). Clustering of gene expression patterns at subsequent steps of B cell development confirmed the similarity between Fr. D resting pre-B cells ex vivo and Ikaros-transduced B3 cells (Figure 6E).
We extended our analysis to primary pre-B cells. As described above for B3 pre-B cells, gene expression changes induced by Ikaros in primary pre-B cells showed strong enrichment for genes that were differentially expressed during B cell lineage specification (Figure 6F left; supplemental Excel File 5) and during the Fr.C' to Fr.D transition (Figure 6F right; supplementary Excel File 5). We conclude that Ikaros target genes are differentially expressed in the context of hematopoiesis, lymphocyte lineage choice, and B cell differentiation.
Ikaros targets in pre-B cells and genes differentially expressed during lymphoid lineage specification and differentiation in vivo share biological pathways
We analyzed the contribution of Ikaros to biological pathways defined by gene expression changes at 4 key stages of differentiation: differentiation of multipotent to lymphoid-restricted progenitors (MLP to CLP), B-cell lineage specification (CLP to Fr.A), T-cell lineage specification (CLP to DP thymocytes), and the transition from cycling to resting pre-B cells (Fr.C’ to D). Gene sets that are up-regulated (Figure 7 yellow) or down-regulated (Figure 7 blue) at each developmental transition in vivo were annotated for: 1) the fraction of genes bound and regulated (Figure 7 dark-shaded segments) or regulated only (Figure 7 medium-shaded segments) by Ikaros at 2, 6, or 48 h; and 2) biological pathways enriched at each developmental transition in vivo as well as among genes that are bound and regulated by Ikaros in B3 cells (P < .05). Note that differential expression in vivo and binding and regulation by Ikaros in vitro define similar biological pathways, especially for gene sets with a large share of Ikaros targets, in particular MLP to CLP up-regulated genes (immune system functions, signaling, activation, proliferation, differentiation, and V(D)J recombination), CLP to Fr.A up-regulated genes (cell cycle and activation), CLP to T-cell progenitor down-regulated genes (metabolic process, ribosome biogenesis, translation, and DNA replication), Fr.C' to D up-regulated genes (immune system, signaling, activation, differentiation, and transcription), and Fr.C' to D down-regulated genes (cell cycle, DNA replication, ribosome biogenesis and translation, nucleoside metabolism, DNA recombination, and repair) (Figure 7).
Ikaros targets in pre-B cells and genes differentially expressed during lymphoid lineage specification and differentiation in vivo share biological pathways. Circles represent up- and down-regulated genes during the in vivo differentiation of MLPs to CLPs, B-cell lineage specification (CLP to Fr.A pro-B cells), T-cell lineage differentiation (CLP to DP thymocytes), and the transition from proliferating to resting pre-B cells (Fr. C' to Fr. D; www.immgen.org). The proportion of these in vivo gene sets that are regulated (intermediate shading) or bound and regulated by Ikaros in B3 cells (dark shading) is shown for the indicated time points. Enriched biological pathways (P < .05) were derived from genes that are differentially expressed in vivo and that are bound and regulated by Ikaros in B3 cells (P < .05). The network of Ikaros-interacting transcription factors depicted at the center of the scheme is based on the literature17,18,33,45 and data described here.
Ikaros targets in pre-B cells and genes differentially expressed during lymphoid lineage specification and differentiation in vivo share biological pathways. Circles represent up- and down-regulated genes during the in vivo differentiation of MLPs to CLPs, B-cell lineage specification (CLP to Fr.A pro-B cells), T-cell lineage differentiation (CLP to DP thymocytes), and the transition from proliferating to resting pre-B cells (Fr. C' to Fr. D; www.immgen.org). The proportion of these in vivo gene sets that are regulated (intermediate shading) or bound and regulated by Ikaros in B3 cells (dark shading) is shown for the indicated time points. Enriched biological pathways (P < .05) were derived from genes that are differentially expressed in vivo and that are bound and regulated by Ikaros in B3 cells (P < .05). The network of Ikaros-interacting transcription factors depicted at the center of the scheme is based on the literature17,18,33,45 and data described here.
Discussion
Although numerous previous studies had focused on individual Ikaros targets in B-cell progenitors, the global identification of Ikaros targets presented here allowed us to address aspects of the biology of Ikaros in developing B cells that were not previously accessible.
First, we were able to evaluate the involvement of Ikaros-regulated genes in key aspects of B-cell progenitor biology, including pre-BCR signaling, cell cycle regulation, and the somatic rearrangement of Ig genes. The temporal resolution of the inducible Ikaros system allowed us for the first time to identify cell cycle genes regulated by Ikaros at time points before Ikaros expression had resulted in G1 arrest. Myc, Cdk6, Ccnd2, Cdkn1a, and Cdkn1b were significantly regulated prior to cell cycle arrest.
Second, we addressed the contribution of Ikaros to the specification of the B-cell lineage and the subsequent differentiation of B-cell progenitors. We achieved this by integrating genome-wide Ikaros targets with gene expression changes that occur between discrete stages of differentiation in vivo. Ikaros targets accounted for a large proportion (50%) of genes up-regulated during B-cell lineage specification in vivo, providing a potential explanation for the essential role of Ikaros in this process. Remarkably, inducible Ikaros expression in cycling pre-B cells was sufficient to drive transcriptional changes that resembled the differentiation of cycling to resting pre-B cells in vivo. This suggests that the incremental expression of the Ikaros paralogs Ikzf1 and Ikzf3 has an important role in driving the differentiation of B-cell progenitors. Ikzf3-deficient pre-B cells display defective silencing of the Ikaros target Igll1 in vivo,13 and genome-wide expression profiling41 showed that ∼50% of transcripts deregulated in Ikzf3-deficient pre-B cells (adjusted P value <0.05) were found on our list of Ikaros-regulated genes, including Igll1, Vpreb1, Ccnd2, Lmo2, Lef1, and Socs2 (not shown). Although Aiolos regulated the expression of all Ikaros targets we have tested, the precise in vivo roles of each paralog remain to be addressed.
Metazoan gene expression networks are both robust and adaptable to withstand intrinsic and extrinsic noise while allowing cellular differentiation to progress. Whereas some network nodes are highly buffered and confer robustness, others act as driver nodes that are critical for changing the network configuration.42 The expression of Ikaros family members responds to pre-BCR/SLP65 signals,13 IRF4,17 and other inputs. It drives developmental progression from the proliferative to the resting stages of pre-B–cell differentiation. Ikaros is necessary for B-cell development but not sufficient to specify B-cell fate (cells of other hematopoietic lineages also express Ikaros family members). Ikaros is therefore not a master regulator in the sense of conferring lineage identity, but rather a driver, which moves this biological system along a predefined trajectory. We suggest that Ikaros dosage acts as a driver that reconfigures the gene expression network in B-cell progenitors from a state resembling cycling Fr.C’ pre-B cells to a state resembling resting Fr.D pre-B cells. These considerations may have implications for addressing the loss of Ikaros in hematopoietic malignancies. First, unlike network hubs, driver nodes can have limited connectivity42 and it may therefore be possible to redress network deviations that result from mutation or loss of Ikaros by manipulating a limited number of key targets. Second, alterations in the gene expression network that result from mutation or loss of Ikaros may sensitize cells to manipulations that are well tolerated by the original network state, suggesting the feasibility of systematic approaches to explore the vulnerabilities of Ikaros-deficient cells.43,44 Third, exploration of the regulatory inputs that determine the expression of individual Ikaros family members might enable approaches to compensate for mutation or loss of Ikaros by other members of the family. Finally, it will be interesting to use the approaches described here to explore the ability of other transcription factors to act as drivers of differentiation in a range of developmental systems.
In summary, genome-wide mapping of Ikaros targets has shown that Ikaros-regulated genes are highly represented in pre-BCR signaling, cell cycle regulation, and the somatic rearrangement of Ig genes, which are key to the differentiation of B-cell progenitors. Integration of Ikaros targets with developmentally regulated gene expression changes provides a rationale for the essential role of Ikaros transcription factor family in the specification and differentiation of B-cell progenitors. We suggest that this approach can be applied to map the contribution of individual transcription factors, transcription factor families, and, potentially, groups of transcription factors to cell lineage commitment and differentiation in other developmental systems.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs. A. Uren and A. Polleri for valuable discussion, Dr. Seden Grippon for help with early stages of the project, Drs. L. Game and M. Jones for high throughput sequencing, A. Giess for sequence alignment, and Drs. J. Elliott and P. Hexley for cell sorting.
This work was supported by the Medical Research Council, UK, Lymphoma and Leukaemia Research (I.F.-V.), the Spanish Ministry of Education (I.F.-V.), and a Chinese Excellence Award (Z.W.L.).
The data in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession numbers GSE38110 (gene expression data for B3 cells), GSE42551 (gene expression data for primary pre-B cells), GSE38169 (ChIP-seq data for B3 cells), and GSE42462 (ChIP-seq data for primary pre-B cells).
Authorship
Contribution: I.F.-V., B.T., Z.W.L., and B.S.C. designed and conducted the experiments; T.C., A.T., G.D., and P.S. analyzed data; I.F.-V., B.T., Z.W.L., B.S.C., T.C., A.T., G.D., P.S., L.B., S.T.S., M.S., E.P., A.G.F., and M.M. contributed to the project outline; and M.M. wrote the manuscript.
Conflict-of-Interest disclosure: The authors declare no competing financial interests.
The current affiliation for B.T. is MRC Laboratory of Molecular Biology, Hills Road, Cambridge CB2 0QH, United Kingdom.
The current affiliation for T.C. is Cancer Research UK, Cambridge Research Institute, Cambridge CB2 1QR, United Kingdom.
The current affiliation for B.S.C. is The Royal Veterinary College, London NW1 0TU, United Kingdom.
The current affiliation for A.T. is Queen Mary, University of London, London E1 4NS, United Kingdom.
Correspondence: Matthias Merkenschlager, Imperial College London, Hammersmith Hospital Campus, Du Cane Road, London, W12 0NN, United Kingdom; e-mail: matthias.merkenschlager@csc.mrc.ac.uk.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal