Abstract
We demonstrate a methodology for tracing the clonal history of hematopoietic stem and progenitor cells (HSPCs) behavior in live tissues in 4 dimensions (4D). This integrates genetic combinatorial marking using lentiviral vectors encoding various fluorescent proteins (FPs) with advanced imaging methods. Five FPs: Cerulean, EGFP, Venus, tdTomato, and mCherry were concurrently used to create a diverse palette of color-marked cells. A key advantage of imaging using a confocal/2-photon hybrid microscopy approach is the simultaneous assessment of uniquely 5FP-marked cells in conjunction with structural components of the tissues at high resolution. Volumetric analyses revealed that spectrally coded HSPC-derived cells can be detected noninvasively in various intact tissues, including the bone marrow, for extensive periods of time after transplantation. Live studies combining video-rate multiphoton and confocal imaging in 4D demonstrate the possibility of dynamic cellular and clonal tracking in a quantitative manner. This methodology has applications in the understanding of clonal architecture in normal and perturbed hematopoiesis.
Introduction
Modern microscopy is an invaluable tool, with advances in resolution, contrast, molecular specificity, speed, and biocompatibility to enable visualization of cellular processes in intact tissues and organisms. The use of endogenously produced multicolor fluorescent proteins (FPs) to label cells has emerged as a versatile approach for cell tracking and lineage tracing during morphogenesis or regenerative processes.1-3 However, many FP variants have similar excitation and emission properties, making unambiguous separation of signals from multiple reporters challenging. The ability to label with multiple FPs in the same experiment and apply high resolution multidimensional imaging can provide insights into complex biologic processes.4
Recently, the concept of genetically labeling clonal cell populations via fluorescent proteins of distinct colors has been developed. The initial technology known as the “brainbow”5 was based on controlled transgene recombination, and successive improvements increased the range of applications.6-9 Up to 5 different FP were expressed from a single “MultiLabel” expression plasmid using tandem recombineering induced by a tissue- and stage-specific promoter, resulting in homogeneous cell populations all expressing multiple FPs.10,11 To date, this approach has not been applied to hematopoiesis because of the lack of a highly hematopoietic stem cell-specific promoter. An alternative strategy, known as “RGB marking” uses lentiviral gene ontology (LeGO) vectors encoding red, green, and blue FPs to transduce multiple cell types and track clones after transfer,12-14 resulting in fluorescence intensities much brighter than most transgenic FPs, and broad combinatorial color diversity.
We took advantage of LeGO vectors constitutively expressing 5FPs to mark hematopoietic stem and progenitor cells (HSPCs) and study the process of hematopoiesis at a clonal level over time and in multiple tissues. HSPCs reside within the BM in a complex niche consisting of osteoblasts, stromal cells, adipose tissue, and vascular structures, crucial for maintenance of self-renewal and modulation of differentiation and death pathways.15-18 As BM has been inaccessible to direct observation, the interactions between HSPCs and their microenvironment remains largely uncharacterized in vivo. Recently, we established a methodology to visualize the 3D architecture of intact BM using confocal fluorescence and reflection microscopy.19
We combine generation of a diverse palette of clone colors via cotransduction of HSPCs with 5FPs LeGO vectors with new imaging and analysis technologies to computationally reconstruct the 3D architecture of tissues at high resolution to depths of 150-300 μm, elucidating biologically interesting clonal reconstitution patterns. We demonstrate that confocal imaging can be combined with multiphoton microscopy, revealing complementary information from autofluorescent and second-harmonic-generating (SHG) structures. Furthermore, dynamic 4D high-resolution imaging is achievable by using video-rate scanning, red-shifted FPs, and longer wavelengths lasers.
Methods
Lentiviral vector production, titration, and cell line studies
The LeGO plasmids expressing FPs from a strong constitutive internal viral promoter, including LeGO-Cer2 (Cerulean), LeGO-G2 (EGFP), LeGO-V2 (Venus), LeGO-T2 (tdTomato), and LeGO-C2 (mCherry), were provided by Boris Fehse (Hamburg, Germany).12 LeGO vectors pseudotyped with VSV-G envelope were produced via cotransfection of 293T cells with a LeGO plasmid concurrently with pCDNA3.HIVgag/pol.4xCTE, pMD2.G-VSV-G, and pRev-TAT.20 Viral supernatants were concentrated via ultracentrifugation, and titers determined on NIH3T3 cells.21 NIH3T3 cells were transduced once at an MOI of 0.5-1 to achieve 50% expression efficiency for each FP. Cells were transduced with each LeGO vector individually and then mixed after transduction (termed Mix) or cotransduced with all 5 LeGO vectors simultaneously (termed Co).
Mouse cell collection, purification, transduction, and transplantation
All mouse procedures were performed with approval from the National Heart, Lung, and Blood Institute Animal Care and Use Committee. Female B6.SJL-Ptprc(d)Pep3(b)/BoyJ (B6.SJL) and C57Bl/6 Ly5.2 mice (The Jackson Laboratory), 6-16 weeks old, were used as donor and recipient, respectively. Marrow was flushed from humeri, femurs, and tibias, red cells were lysed with ACK buffer (Quality Biologic), and the MACS lineage depletion kit (Miltenyi Biotec) was used to purify lineage negative (Lin−) progenitor cells.
Lin− cells were cultured for 48 hours at a starting density of 5 × 105 cells/mL in StemSpan media (StemCell Technologies) supplemented with 10 ng/mL murine IL-3, 100 ng/mL murine IL-11, 100 ng/mL human Flt-3 Ligand (all from R&D Systems) and 50 ng/mL murine stem cell factor (Research Diagnostics). Cells were transferred to dishes coated with RetroNectin (Takara) and transduced with LeGO supernatants each at a MOI of 6-7 in the presence of the media and cytokines detailed above, and 4 μg/mL protamine sulfate (Sigma-Aldrich). After 24 hours, the cells were removed from culture for transplantion or transferred to fresh media and cultured for an additional 96 hours before flow cytometric analysis, plating in a CFU assay, or confocal microscopy. CFU assays were carried out according to the manufacturer's instructions (StemCell Technologies) using a starting number of 1000 Lin− cells.
Transduced Lin− HSPCs were infused by tail vein injection into 11-Gy–irradiated recipients, at doses corresponding to 1-4 × 105 Lin− cells per mouse, quantitated at the time the cells were placed into culture. A total of 5 independent experiments were performed: 2 with Co 5FPs, one with Mix 5FPs, one with Co 3FPs, and one with single FPs. In each experiment, cohorts of animals were transplanted with the same population of transduced donor BM cells, and individual mice were killed for tissue retrieval and imaging at various time points up to 120 days after transplantation.
To perform secondary transplants, whole BM nucleated cells from 2 primary Mix 5FPs mice were transplanted using a ratio of 1 primary mouse to 4 secondary mice, into 11-Gy–irradiated secondary C57/Bl6 (Ly5.2) recipients.
For imaging, spleen, popliteal lymph nodes, lung, heart, skin, adipose tissue, skeletal muscle, kidney, and liver were excised and imaged intact without sectioning, fixation, or further processing. Calvarial marrow was imaged directly through the skull without sectioning. Sternal marrow was imaged after bisection with a razor along the sagittal plane.19,22-24
Flow cytometric analyses
The transduction efficiency for NIH3T3 cells or Lin− cells was assessed using a BD Fortessa LsrII flow cytometer (BD Biosciences).
Confocal and 2-photon microscopy
Microscopy imaging was performed using a Leica TCS SP5-AOBS 5-channel confocal and multiphoton system (Leica Microsystems) equipped with multiline Argon, diode 561-nm, HeNe 594-nm, and HeNe 633-nm visible lasers.
The confocal spectral mode (xyλ) was used to collect emitted light in 5-nm bandwidths of the visible spectrum from NIH3T3 cells transduced with a single FP LeGO vector. Spectral images were recorded using several laser excitations wavelengths: Cerulean (458 and 488 nm), EGFP (488 and 514 nm) Venus (488 and 514 nm), tdTomato (561 and 514 nm), and mCherry (561 and 594 nm). Images were processed with the Leica LAS-AF Version 2.4.1 software to create reference spectra for all 5FPs. Reference emission spectra were recorded also from Lin− transduced with single FP-encoding vectors and from mouse tissues of animals transplanted with single FP-transduced cells and nontransplanted controls. Using the individual FP spectra and the difference in brightness, 5 sequential confocal imaging channels were set to exclusively capture the corresponding FP without crosstalk. Thus, each marked cell has a unique spectral identity characterized in 5-channel (8-bits) color information. For each FP, fluorescence intensity profiles were analyzed along lines drawn manually through the center of a cell (in Imaris software) to distinguish cells in an unbiased manner, independent of visual inspection.
In 2-photon mode, a pulsed femtosecond Ti:Sapphire (Ti-Sa) Chameleon Vision II laser (Coherent), tunable for excitation from 680 to 1080 nm with dispersion correction, was used. Fluorescence emission was separated by custom dichroic mirrors (Semrock) and collected backward with 4 channels nondescanned detectors (Leica). Wavelength separations were as follows: dichroic 568 nm followed by dichroic 465 nm, followed by a 452/45 nm emission filter for SHG or autofluorescence and Cerulean, a 525/40 nm filter for EGFP or Venus, and a 648 nm dichroic followed by a 620/60 nm filter for tdTomato or mCherry. Absorption spectra of single FP containing tissues and controls were recorded to determine the most favorable 2-photon excitation wavelength.
To reveal structural information, we used 2-photon intrinsic contrast imaging of tissue autofluorescence from elastin, NADH (excited at 780 nm), and second harmonic generated signal (SHG) from bone and fibrillar collagen (excited at 920 nm or 860 nm, collected backward).25-27 In some experiments involving red-shifted FPs (tdTomato and mCherry), an optical parametric oscillator (OPO) laser (Chameleon Compact OPO) from Coherent was used to extend the output wavelength range as far as 1030-1300 nm, sequentially or simultaneously with the Ti-Sa laser.
Images of the tissues were taken using a HCX-IRAPO-L 25×/0.95 NA water dipping objective (WD = 2.5 mm), a HC-PLAPO-CS 20×/0.70 NA dry objective (WD = 0.6 mm), or HC-PL-IRAPO 40×/1.1 NA water immersion objective (WD = 0.6 mm). Images of cultured cells were taken with a 40× 1.4 NA oil immersion objective.
For 3D volume rendering, series of x-y-z images (typically 1 × 1 × 4 μm3 voxel size) were collected along the z-axis at 5-μm intervals throughout the tissues (150-300 μm) depth, over large regions, using the tile function of the software to automatically generate stitched volumes, for instance composing an entire sternal fossae, ∼ 2.5 × 1.2 mm2 (x-y) and 300 μm (z).
For 4D time-lapse imaging, popliteal lymph nodes, spleen, lung, or calvarial bone samples were placed uncut onto 35-mm number 0 cover glass culture dishes (MatTek), in 50-100 μL DMEM, 10% FBS containing 20mM HEPES at 37°C and 2-photon Ti-Sa excitation (860 nm) combined with confocal excitation (458 nm, 488 nm, 514 nm, 561 nm, 594 nm) was performed using a Leica resonant scanner (8000 Hz/s), taking z-stacks (∼ 80 μm) at 20-second intervals for up to 1 hour. In select experiments, Ti-Sa tuned at 860 nm was used simultaneously with the OPO laser (tuned at 1130 nm-for tdTomato; or 1140 nm mCherry) for simultaneous 3-color 2P imaging.
Reconstruction and analysis of 3D volumes and 4D time sequences
The tiled-stacks of raw images were transformed into volume-rendered (3D) data of fluorescent cells using Imaris x64 Versions 7.2 and 7.4 software (Bitplane) and exported as 3D-rotation movies, as described.19 Some images were represented as orthogonal projections (x-y-z directions) of 10-20 optical sections in 5-μm increments (extended depth of focus) rendered using Imaris software. For quantitative assessment of 3D cell and clone frequency, positions, sizes, and distribution, z-stacks were further processed using the Imaris XT modules, which integrate MATLAB applications (MathWorks) by distance measurements (using a distance transformation algorithm) and cluster analysis, as described.19 The sequences of image stacks over time were transformed into volume-rendered 4-dimensional (4D) movies with Imaris x64 software, and spot and surface analysis was used for semiautomated tracking of cell motility in 3D using autoregressive motion algorithms. Multicolor labeled spots were extracted by combining all channels; spots < 2 μm apart were linked to account for cells labeled with more than one color and tracked over time. The dataset was corrected for tissue drift, and calculations of the cell velocity and displacement over time, track lengths, paths, and distances were performed; values were exported in Excel software (Microsoft) to plot graphs. Composite figures were assembled with Adobe Photoshop CSS 5. Movie compression was performed with Adobe Premiere Version 6.5 software.
Results
Imaging 5FPs by confocal microscopy: proof of principle
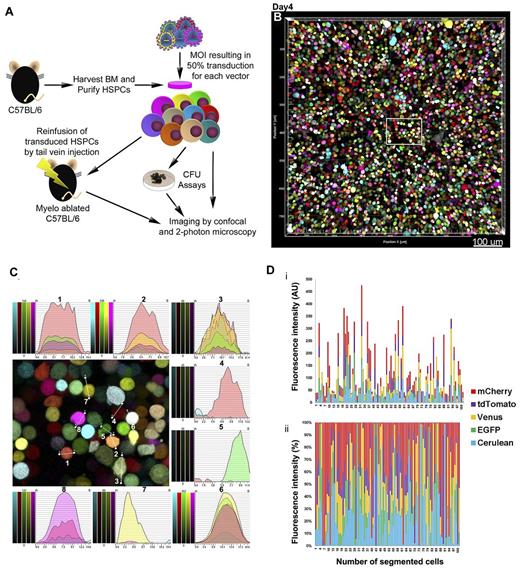
We first established that cells labeled with 5FPs with highly overlapping emission spectra could be distinguished via spectrally acquired confocal microscopy. NIH3T3 fibroblasts transduced with the LeGO vectors encoding the 5FPs were used to test feasibility. Figure 1 illustrates the steps taken: spectral (lambda) xyλ images were acquired for cells transduced individually with each of the 5FP vectors (single FPs) to create reference spectra for each FP (Figure 1A). Unexpectedly, we observed that the mCherry spectrum was dependent on the excitation wavelength, and that 594 nm excitation resulted in ∼ 12-nm-peak red-shift compared with 561-nm excitation. Using the individual FP spectra and difference in brightness (expressed as % of EGFP) of Cerulean (79%), Venus (156%), tdTomato (283%), and mCherry (47%), we were able to define 5 distinct sequential imaging channels collecting in the following emission (em) windows: Cerulean (458 em 468-482), EGFP (488 em 496-514), Venus (514 em 523-558), tdTomato (561 em 579-597), and mCherry (594 em 618-670; Figure 1A-B). This approach resulted in accurate separation, as cells transduced with each individual FP were visible only in the corresponding channel and absent from the rest (Figure 1C). This defined multichannel approach had the advantage of reducing acquisition times compared with spectral imaging across the entire spectrum.
Confocal microscopy approach for imaging cells transduced with LeGO vectors encoding 5FPs. NIH3T3 cells transduced with LeGO vectors encoding single FPs were used to define imaging parameters. (A) Spectral (xyλ) imaging was used to record reference emission spectra for each FP, illustrated in normalized histograms pseudo-colored with cyan (Cerulean), green (EGFP), yellow (Venus), magenta (tdTomato), and red (mCherry). The mCherry spectrum red shifts (12 nm at peak) when excited by 594 nm (solid red line) compared with 561 nm (dashed red line) wavelength. (B) Five channels were set to image sequentially: Cerulean (458 emission [em] 468-482), EGFP (488 em 496-514), Venus (514 em 523-558), tdTomato (561 em 579-597), and mCherry (594 em 618-670). (C) Imaging of NIH3T3 cells transduced with individual FP vectors. Each FP was visible only in the cells transduced with the corresponding vector imaged in the appropriate channel, and absent from the others (no crosstalk). (D-E) Populations of NIH3T3 cells transduced independently with the 5 LeGO vectors and then mixed (Mix, D) or NIH3T3 cells transduced with all 5 vectors concurrently (Co, E), plated and imaged 4-5 days later. (D) The 5 individual FPs are shown pseudo-colored cyan (Cerulean), green (EGFP), yellow (Venus), magenta (tdTomato), and red (mCherry). (E) Clusters of progeny cells inheriting a specific color within a diverse palette of colors based on combinations of 1-5 FPs at variable intensities in each originally transduced cell. Progeny inherit the specific color, allowing tracking of lineage relationships (related images; supplemental Videos 1 and 2). Scale bar represents 100 μm (D-E).
Confocal microscopy approach for imaging cells transduced with LeGO vectors encoding 5FPs. NIH3T3 cells transduced with LeGO vectors encoding single FPs were used to define imaging parameters. (A) Spectral (xyλ) imaging was used to record reference emission spectra for each FP, illustrated in normalized histograms pseudo-colored with cyan (Cerulean), green (EGFP), yellow (Venus), magenta (tdTomato), and red (mCherry). The mCherry spectrum red shifts (12 nm at peak) when excited by 594 nm (solid red line) compared with 561 nm (dashed red line) wavelength. (B) Five channels were set to image sequentially: Cerulean (458 emission [em] 468-482), EGFP (488 em 496-514), Venus (514 em 523-558), tdTomato (561 em 579-597), and mCherry (594 em 618-670). (C) Imaging of NIH3T3 cells transduced with individual FP vectors. Each FP was visible only in the cells transduced with the corresponding vector imaged in the appropriate channel, and absent from the others (no crosstalk). (D-E) Populations of NIH3T3 cells transduced independently with the 5 LeGO vectors and then mixed (Mix, D) or NIH3T3 cells transduced with all 5 vectors concurrently (Co, E), plated and imaged 4-5 days later. (D) The 5 individual FPs are shown pseudo-colored cyan (Cerulean), green (EGFP), yellow (Venus), magenta (tdTomato), and red (mCherry). (E) Clusters of progeny cells inheriting a specific color within a diverse palette of colors based on combinations of 1-5 FPs at variable intensities in each originally transduced cell. Progeny inherit the specific color, allowing tracking of lineage relationships (related images; supplemental Videos 1 and 2). Scale bar represents 100 μm (D-E).
We next imaged NIH3T3 cell populations transduced with the 5FP LeGO vectors individually at an MOI of 0.5-1 and then mixed before imaging (Mix) or NIH3T3 cells transduced with all 5FP vectors concurrently (Co), at an MOI predicted to result in the greatest diversity of FP combinations between individual transduced cells, comparing our multichannel approach with spectral imaging followed by linear unmixing. Figure 1D shows the merged 5-channel images of the Mix cell population, delineating clonal clusters of NIH3T3 cells expressing one of the 5 individual FPs (supplemental Video 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Figure 1E shows the 5 merged channel image of the Co cell population, demonstrating clusters of cells expressing a highly diverse palette of colors resulting from the combination of signals of 1-5 different FPs at variable intensities, probably depending on the number of copies and the particular subset of vectors transducing each cell. Each pixel in the image has a unique spectral identity characterized by 5-channel (8 bits/channel) intensity values. A 3D collection over a large area is shown in supplemental Video 2.
Application of confocal 5FP imaging to murine HSPCs
To adapt this approach to tracking hematopoietic progeny from HSPCs, we used the same acquisition and image-processing protocols to study primary murine BM cells transduced with the 5FP LeGO vectors. Murine Lin− BM cells were Co-transduced at an MOI (6-7 for each vector) resulting in ∼ 50% transduction efficiency by each vector individually, as assessed by flow cytometry, and imaged 96 hours later (Figure 2A). We were able to image over large fields of view at single-cell resolution in 3D (Figure 2B-C; supplemental Video 3), demonstrating individual cells displaying highly diverse color marking, resulting from transduction of each HPSC with a different selection and number of LeGO vectors. Fluorescence line intensity profile analyses demonstrate unique spectral identities, independently of visual color inspection (Figure 2C). Furthermore, cells can be automatically segmented as individual spots objects with characteristic mean fluorescence intensity in each of the 5 channels creating an objective tool to distinguish them for further spatial-temporal analyses (Figure 2D; supplemental Table 1).
Confocal microscopy imaging of primary BM HSPCs cotransduced with 5FP lentiviral vectors. (A) A schematic overview of the transplantation of LeGO-transduced murine HSPCs. Donor Lin− HSPCs were harvested, transduced in vitro with all 5FP vectors concurrently (Co) as shown in the schematic, or individually and then mixed (Mix), and transplanted into lethally irradiated recipient mice or continued in culture for flow cytometric, CFU, and imaging analyses. Tissues from transplanted recipients were removed at specific time points for imaging. (B) Lin− cells co-transduced with the 5 LeGO vectors and imaged 4 days after transduction, showing a merged tiled-3D image of highly diverse color marking of individual cells, resulting from the variety of vector combinations transducing each cell. (C) Higher magnification view of the boxed area from the image shown in panel B; line intensity profiles were drawn manually through the center of 8 cells, generating 5-channel intensity histograms demonstrating unique spectral “colorprints” to unequivocally distinguish each transduced cell. (D) Cells from panel B were segmented as spot objects using Imaris software, and the MFI/spot in each of the 5 channels was quantitated and displayed as stacked columns, either as (i) absolute values (AU arbitrary units) or as (ii) percent relative contributions of the 5 FP channels. This graphically and quantitatively demonstrates the huge diversity of spectral identity for individual Lin− cells. Original raw data for the first 100 spots are shown in supplemental Table 1. Scale bar represents 100 μm.
Confocal microscopy imaging of primary BM HSPCs cotransduced with 5FP lentiviral vectors. (A) A schematic overview of the transplantation of LeGO-transduced murine HSPCs. Donor Lin− HSPCs were harvested, transduced in vitro with all 5FP vectors concurrently (Co) as shown in the schematic, or individually and then mixed (Mix), and transplanted into lethally irradiated recipient mice or continued in culture for flow cytometric, CFU, and imaging analyses. Tissues from transplanted recipients were removed at specific time points for imaging. (B) Lin− cells co-transduced with the 5 LeGO vectors and imaged 4 days after transduction, showing a merged tiled-3D image of highly diverse color marking of individual cells, resulting from the variety of vector combinations transducing each cell. (C) Higher magnification view of the boxed area from the image shown in panel B; line intensity profiles were drawn manually through the center of 8 cells, generating 5-channel intensity histograms demonstrating unique spectral “colorprints” to unequivocally distinguish each transduced cell. (D) Cells from panel B were segmented as spot objects using Imaris software, and the MFI/spot in each of the 5 channels was quantitated and displayed as stacked columns, either as (i) absolute values (AU arbitrary units) or as (ii) percent relative contributions of the 5 FP channels. This graphically and quantitatively demonstrates the huge diversity of spectral identity for individual Lin− cells. Original raw data for the first 100 spots are shown in supplemental Table 1. Scale bar represents 100 μm.
The transduced cells were plated in semisolid methylcellulose media to generate clonal colony-forming units (CFU), with each CFU expressing one of a diverse palette of colors and all cells in each individual clonal CFU colony having the same spectral fingerprint generated by the 5-channel image collection and reconstruction (supplemental Figure 1A-C; supplemental Table 1). We illustrate individual CFUs marked by up to 5FPs merged and as 5 individual channel images. Single-cell quantitative image analysis reveals homogeneous FP relative expression levels between individual cells in the same colony.
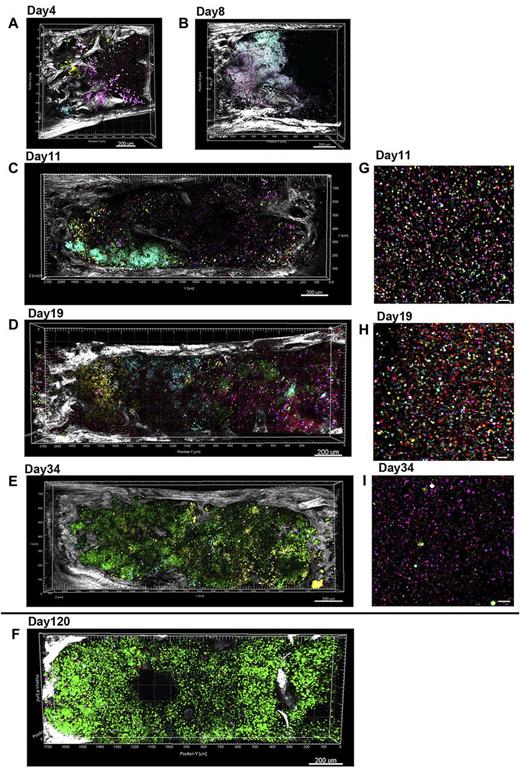
We performed BM transplants with 5FP Mix or Co-transduced HSPCs into lethally irradiated recipient mice, and followed regeneration of hematopoiesis in the spleen and BM over time, via confocal combined with 2-photon (SHG) microscopy (Figure 3). In addition to intracellular fluorescence, 2-photon imaging can also detect second harmonic signals, generated when 2-photons of a given wavelength are absorbed and replaced by a single photon of half the wavelength. This process does not require exogenous labeling and is primarily used to visualize collagen networks.28 This combination imaging allows clear visualization of collagen-rich bone structure and the interaction between HSPC clones and their microenvironment. Cohorts of animals were transplanted with the same population of transduced donor Lin− cells, and individual mice were killed at various time points, including 4, 8, 11, 19, 34, and 120 days after transplantation, to retrieve organs for imaging. The best-preserved and highest resolution imaging of large volumes of regenerating BM was obtained using whole mounts of bisected sternums. We visualized active polyclonal engraftment with clones of diverse colors occurring in close proximity to bone very early after transplantation (day 4 and 8 after transplantation, Figure 3A-B; supplemental Video 4). Over time, the individual clones expanded and began to occupy more central marrow regions (days 11-19, Figure 3C-D). By day 34 and later, interestingly large areas of the sternum were occupied by individual, mostly monochromatic noninterspersed clones, as shown in Figure 3E and F and supplemental Video 5. Imaging of peripheral blood leukocytes at the same time points (Figure 3G-I) revealed a similar pattern of highly polyclonal early hematopoiesis, progressing to a more oligoclonal pattern. But individual hematopoietic clones remained geographically demarcated in the marrow even at late time points. We also examined calvarial (skull) BM around central veins and coronal sutures, over large volumes, without sectioning, and observed similar clonal clusters of cells in apposition to bone (supplemental Figure 2; supplemental Video 6).
Engraftment and expansion of transplanted 5FP-marked cells in the BM in vivo. Lin− cells transduced with 5FPs were infused into mice after total body irradiation. Bisected whole sternal mounts were examined at different time points by confocal and 2-photon microscopy (A-F). Peripheral blood nucleated cells from the same time point are shown to the right of each sternal marrow image (G-I). For all images, tiled z-stacks were collected over large volume (2.5-mm × 1.2-mm × 300-μm) of the mouse BM tissue and computationally stitched. (A-E) Different mice, each receiving the same population of Co 5FP–transduced donor cells. (F) Different experiment of Mix 5FP–transduced donor cells. Related images are found in supplemental Videos 4 and 5. (A-B) Endosteal engraftment occurring early in BM regeneration (day 4 and 8 after transplantation, respectively); clusters of cells marked in wide variety of colors were visible in close proximity to the bone edge (SHG, white). (C) By day 11, multiple cell clusters are located in close proximity to the bone and progressing centrally over time. (D) By day 19, multiple uniquely color-marked clones are distinguishable over the entire marrow fossae. (E) By day 34, one large clone (green) has expanded to occupy the entire fossae. However, the peripheral blood from the same time point (I) continues to show the presence of multiple clones contributing to hematopoiesis. (F) Day 120 sternum is shown from another experiment, with animals receiving Mix 5FP–transduced BM, similarly showing a very large single clone occupying most of the sternal marrow space. Scale bars represent 200 μm (A-F), 50 μm (G-H).
Engraftment and expansion of transplanted 5FP-marked cells in the BM in vivo. Lin− cells transduced with 5FPs were infused into mice after total body irradiation. Bisected whole sternal mounts were examined at different time points by confocal and 2-photon microscopy (A-F). Peripheral blood nucleated cells from the same time point are shown to the right of each sternal marrow image (G-I). For all images, tiled z-stacks were collected over large volume (2.5-mm × 1.2-mm × 300-μm) of the mouse BM tissue and computationally stitched. (A-E) Different mice, each receiving the same population of Co 5FP–transduced donor cells. (F) Different experiment of Mix 5FP–transduced donor cells. Related images are found in supplemental Videos 4 and 5. (A-B) Endosteal engraftment occurring early in BM regeneration (day 4 and 8 after transplantation, respectively); clusters of cells marked in wide variety of colors were visible in close proximity to the bone edge (SHG, white). (C) By day 11, multiple cell clusters are located in close proximity to the bone and progressing centrally over time. (D) By day 19, multiple uniquely color-marked clones are distinguishable over the entire marrow fossae. (E) By day 34, one large clone (green) has expanded to occupy the entire fossae. However, the peripheral blood from the same time point (I) continues to show the presence of multiple clones contributing to hematopoiesis. (F) Day 120 sternum is shown from another experiment, with animals receiving Mix 5FP–transduced BM, similarly showing a very large single clone occupying most of the sternal marrow space. Scale bars represent 200 μm (A-F), 50 μm (G-H).
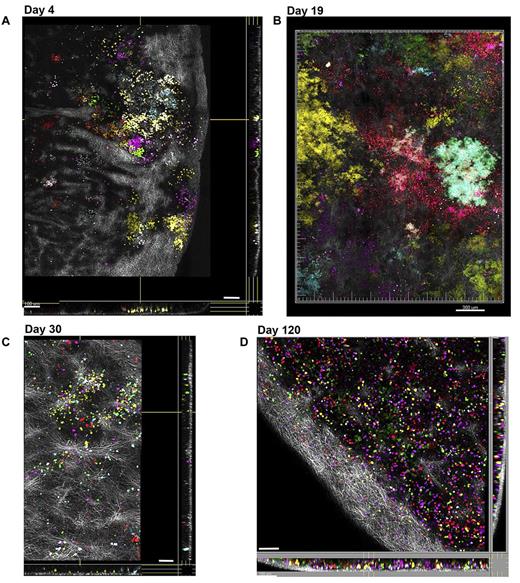
We imaged intact spleens from these animals at the same time points. In the mouse, the spleen is known to be an active site of multilineage hematopoiesis, particularly under stress and early after hematopoietic transplantation. As shown in Figure 4A, at day 4, a great diversity of individually colored hematopoietic clones can be appreciated under the collagen-rich capsule (SHG) at various depths up to 100 μm (supplemental Video 7). By day 19 (Figure 4B; supplemental Figure 3A-C), these color-delineated clonal clusters markedly increased in size to ∼ 500-600 μm (x-y), forming large nodules deforming the spleen surface and corresponding to the morphologically described colony-forming unit spleen (CFU-S) first described by McCullough and Till.29 Each clone consisted of homogeneously colored cells as demonstrated by spectral fingerprinting, with overlap of cells from various clones at the edges (supplemental Figure 3B). A variety of 1, 2, and 3 FPs could be found in the same cells creating clearly distinguishable clones. By day 30 (Figure 4C), when transplanted mice have normalized their blood counts and are fully engrafted in the BM, ongoing splenic clonal hematopoiesis from individual HSPCs is subsiding, and only small clusters of diverse colors are seen, enmeshed by collagen fibers. By day 120 (Figure 4D), all that remains are multiple individual cells derived from diverse clones, most like newly developed T and B cells that have migrated to the spleen from the thymus and BM.
Imaging hematopoietic progenitor clonal output in the spleen over time. In a single experiment, Co 5FP Lin− BM cells were transplanted into multiple recipient mice after total body irradiation. Intact spleens were removed at each time point, imaged over large volumes (1.2-mm × 1.6-mm × 100-μm), and are presented as extended focus sections across x, y, and z dimensions; the superficial layers of the capsule were removed computationally, facilitating visibility of the fluorescent cells underneath. (A) At day 4, numerous small clusters of diverse colors are visible under the collagen-rich capsule (SHG, white). (B) At day 19, very large clonal colonies or CFU-S diameter 500-600 μm (x-y) are present, with color homogeneity associated with each CFU, although by this time point there was clonal mixing at the edges of neighboring CFU-S. (C) At day 30, the large CFU-S had disappeared, with only small residual clusters of clonal cells enmeshed by collagen fibers. (D) At day 120, the spleen is repopulated by scattered nonclonal cells indicated by the color diversity and lack of clustering of cells of the same color. Scale bars represent 100 μm (A,C-D), 300 μm (B). Related images shown in supplemental Video 7.
Imaging hematopoietic progenitor clonal output in the spleen over time. In a single experiment, Co 5FP Lin− BM cells were transplanted into multiple recipient mice after total body irradiation. Intact spleens were removed at each time point, imaged over large volumes (1.2-mm × 1.6-mm × 100-μm), and are presented as extended focus sections across x, y, and z dimensions; the superficial layers of the capsule were removed computationally, facilitating visibility of the fluorescent cells underneath. (A) At day 4, numerous small clusters of diverse colors are visible under the collagen-rich capsule (SHG, white). (B) At day 19, very large clonal colonies or CFU-S diameter 500-600 μm (x-y) are present, with color homogeneity associated with each CFU, although by this time point there was clonal mixing at the edges of neighboring CFU-S. (C) At day 30, the large CFU-S had disappeared, with only small residual clusters of clonal cells enmeshed by collagen fibers. (D) At day 120, the spleen is repopulated by scattered nonclonal cells indicated by the color diversity and lack of clustering of cells of the same color. Scale bars represent 100 μm (A,C-D), 300 μm (B). Related images shown in supplemental Video 7.
To assess how robust the color marking is in vivo, we performed secondary BM transplant experiments (supplemental Figure 4). Total BM donor cells from 2 Mix 5FP primary mice 120 days after transplantation were pooled and transplanted into irradiated recipient mice, and individual mice were killed at various time points, including 5, 20, and 55 days after transplantation. In agreement with primary transplant input, we observed similar, clearly marked, clones of diverse colors in the spleen and sternal BM at day 5 after transplantation. Over time, at day 20 and 55, the majority of cells in the spleen, sternal BM, and peripheral blood still showed cells expressing various FPs, demonstrating the stability of HSPC FP marking over the long-term.
In contrast to the spleen, peripheral lymph nodes in the mouse are not known to be a site for ongoing hematopoiesis, even under stress or during posttransplantation reconstitution. Popliteal lymph nodes were imaged intact, and volumetric images analyzed, reconstructing the node surface area to depths of ∼ 150 μm (supplemental Figure 5). Only rare fluorescent cells were detected in the first 2 weeks after transplantation; by day 19 (supplemental Figure 5A), numerous single cells of diverse colors have begun to enter the node, continuing to increase in number by day 34 and day 120 (supplemental Figure 5B-C; supplemental Video 8).
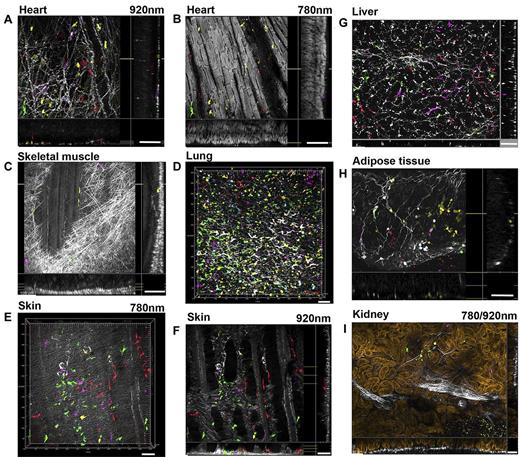
Given intense interest in the potential contributions of BM-derived cells to nonhematopoietic tissues and controversies generated by use of tissue sections to attempt to colocalize marrow-derived FPs or antigens with diverse nonhematopoietic cell types,30,31 we applied our high-resolution 3D imaging to analyze multiple organs to depths of 80-100 μm from mice engrafted with 5FP-transduced HSPCs at day 30 after transplantation. The heart was imaged intact from the epicardial side to depths of 80-90 μm. Scattered fluorescent cells of various colors with macrophage-like morphology, derived from multiple different transduced HSPCs, were visible along superficial collagen fibers (SHG; Figure 5A), and also deeper along cardiomyocytes visualized at high resolution by intrinsic 2-photon autofluorescence (Figure 5B; supplemental Video 9). No FP-positive cardiomyocytes were detected. In mandibular skeletal muscle (Figure 5C; supplemental Video 10), imaged uncut, fluorescent cells were visible along collagen fibers superficially as well as along skeletal muscle fibers with characteristic striations visible in the SHG image. In the lung (Figure 5D; supplemental Video 11), numerous cells with a large palette of colors and diverse morphologies (dendritic-like, macrophage-like, some suggestive of type 2 pneumocytes) were visible. Imaging of an abdominal skin flap from a mouse transplanted with Mix 5FP cells was performed both from the epidermal (Figure 5E) and dermal (Figure 5F) sides. Cells of all 5 colors are visible in both images, with diverse morphologies, including dendritic or macrophage-like, lying under elastin fiber autofluorescence (Figure 5E) and along collagen fibers, muscle fibers, vascular structures, and hair follicles (Figure 5F; supplemental Video 12). In the uncut liver (Figure 5G), fluorescent cells with morphologies suggestive of stellate cells or macrophages (Kupffer cells) of various colors were aligned along a collagen fiber network delineating hepatic lobular structures (supplemental Video 13). There were no fluorescent cells resembling hepatocytes. Adipose tissue (Figure 5H) displayed clusters of fluorescent macrophage-like cells infiltrating tissue along collagen fibers outlining the large adipose cells (supplemental Video 14). In the kidney (Figure 5I), examined inward from the capsule, small fluorescent cells were visible along collagen fibers in the interstitial space between tubules, whose structure is defined via 2-photon autofluorescence imaging (supplemental Video 15).
Detection of BM-derived cells in nonhematopoietic tissues after transplantation. Various tissues and organs were imaged intact at day 30 after transplantation of Lin− cells cotransduced with 5FP vectors (A-D,G-I) or Mix 5FP (E-F). Images are presented as extended depth of focus sections in x-y and x-z and y-z directions after computationally removing the outer layers of collagen fibers. In the heart viewed from the epicardial side, (A) numerous individual large fluorescent cells with macrophage-like morphology originating from multiple clones based on the diverse colors seen are visible along the superficial collagen fibers (SHG at 920 nm, white) and also (B) interspersed deeper between cardiomyocytes (white) visualized by their intrinsic 2-photon autofluorescence (at 780 nm). No FP fluorescent cardiomyocytes were observed. In skeletal muscle, (C) fluorescent cells were found within the external fibrous network (SHG, white) but also intercalating between individual striated muscle fibers (SHG, white). In the lung, (D) numerous fluorescent cells with a diverse palette of colors were scattered throughout at all depth levels, with variable morphologies suggesting dendritic cell, macrophage, and type 2 pneumocyte identities. An abdominal skin flap was imaged from the epidermal side (E) and the dermal side (F); fluorescent cells of diverse colors and morphologies were seen, most with large size and morphology suggesting Langerhans or dendritic-like identity, lying under elastin fibers (autofluorescence at 780 nm, white; E) and along collagen (SHG at 920 nm, white) muscle fibers and hair follicles (F). Groups of nearby cells of the same color suggest in situ proliferation of at least some of these cells. In the liver, (G) fluorescent cells with morphology suggestive of stellate cells or macrophages (Kupffer cells) of various colors were aligned along the collagen fiber network (SHG, white) delineating hepatic lobular structures. Nearby cells of the same color suggest in situ proliferation and short-distance migration. Adipose tissue (H) displayed individual cells or clonal clusters of fluorescent macrophage-like cells or small round lymphocyte-like cells interspersed along fibers (SHG, white) and outlining the large adipose cells. In the kidney, (I) examined from the capsule (collagen-SHG at 920 nm, white), rare small fluorescent cells were visible along collagen fibers in the interstitial space between tubules (golden) imaged via 2-photon autofluorescence (780 nm). Scale bars represent 100 μm (A-F), 50 μm (G-I). Related images shown in supplemental Videos 9-15.
Detection of BM-derived cells in nonhematopoietic tissues after transplantation. Various tissues and organs were imaged intact at day 30 after transplantation of Lin− cells cotransduced with 5FP vectors (A-D,G-I) or Mix 5FP (E-F). Images are presented as extended depth of focus sections in x-y and x-z and y-z directions after computationally removing the outer layers of collagen fibers. In the heart viewed from the epicardial side, (A) numerous individual large fluorescent cells with macrophage-like morphology originating from multiple clones based on the diverse colors seen are visible along the superficial collagen fibers (SHG at 920 nm, white) and also (B) interspersed deeper between cardiomyocytes (white) visualized by their intrinsic 2-photon autofluorescence (at 780 nm). No FP fluorescent cardiomyocytes were observed. In skeletal muscle, (C) fluorescent cells were found within the external fibrous network (SHG, white) but also intercalating between individual striated muscle fibers (SHG, white). In the lung, (D) numerous fluorescent cells with a diverse palette of colors were scattered throughout at all depth levels, with variable morphologies suggesting dendritic cell, macrophage, and type 2 pneumocyte identities. An abdominal skin flap was imaged from the epidermal side (E) and the dermal side (F); fluorescent cells of diverse colors and morphologies were seen, most with large size and morphology suggesting Langerhans or dendritic-like identity, lying under elastin fibers (autofluorescence at 780 nm, white; E) and along collagen (SHG at 920 nm, white) muscle fibers and hair follicles (F). Groups of nearby cells of the same color suggest in situ proliferation of at least some of these cells. In the liver, (G) fluorescent cells with morphology suggestive of stellate cells or macrophages (Kupffer cells) of various colors were aligned along the collagen fiber network (SHG, white) delineating hepatic lobular structures. Nearby cells of the same color suggest in situ proliferation and short-distance migration. Adipose tissue (H) displayed individual cells or clonal clusters of fluorescent macrophage-like cells or small round lymphocyte-like cells interspersed along fibers (SHG, white) and outlining the large adipose cells. In the kidney, (I) examined from the capsule (collagen-SHG at 920 nm, white), rare small fluorescent cells were visible along collagen fibers in the interstitial space between tubules (golden) imaged via 2-photon autofluorescence (780 nm). Scale bars represent 100 μm (A-F), 50 μm (G-I). Related images shown in supplemental Videos 9-15.
Reconstruction and analysis of 3D volumes and 4D time sequences
3D renderings described herein, illustrating the density and complexity of BM and various tissues, were software-generated either as static images or animated 3D-rotation movies over large regions of computationally stitched images. To relate clonal cell growth to neighboring structures, we explored the feasibility of quantitative analysis of distribution and association of multicolor-marked clonal cell populations. Using commercially available software segmentation, we were able to computationally extract distinct multicolor clones in the spleen, determine their frequency, and measure their distance from the surface (Figure 6A-C). Although complete automation of segmentation of multicolor-coded cells remains beyond the current state of the art in image analysis software, the software allowed user visual inspection for reviewing and editing the automated results. All 5-channel segmentations and cell classifications in groups can be visualized in context of the 3D renderings, allowing comparison with the original image data and visual validation.
Quantitative analysis of clone size and location in 3D and motility in 4D. Volumetric 3D tiled stacks from the spleen shown in Figure 4A were analyzed in a quantitative manner. Cells were computationally extracted both individually (A) and as 32 clones spectrally distinct from neighboring ones (B) using semiautomatic user-interactive segmentation, combining channels and grouping in Imaris software. Distances of the individual clones to the splenic capsule were determined using distance transformation algorithm of Imaris XT. (C) Histogram depicting the distances of 32 clones to the surface of the spleen. Time sequences recorded in the lymph node 14 days after transplantation of Mix 5FPS Lin− HSPCs using resonant scanner were transformed into 4D movies and analyzed using XT module of Imaris software (supplemental Video 16). (D) Example of cells computationally segmented as “spots” objects and the lymph node capsule as a “surface” object. Cells expressing tdTomato, EGFP, or Cerulean were represented by magenta (M), green (G), or white (W) spots, respectively. (E) Distances of individual cells/spots to the surface were plotted according to the FP expressed (M, G, W) for comparison (the lines show the mean). (F) Cell migration trajectories (tracks) over time and displacement vectors can be analyzed. (G) Distances between individual tracks and their displacement in x,y,z directions reveal cell motility heterogeneity in depth (color coded).
Quantitative analysis of clone size and location in 3D and motility in 4D. Volumetric 3D tiled stacks from the spleen shown in Figure 4A were analyzed in a quantitative manner. Cells were computationally extracted both individually (A) and as 32 clones spectrally distinct from neighboring ones (B) using semiautomatic user-interactive segmentation, combining channels and grouping in Imaris software. Distances of the individual clones to the splenic capsule were determined using distance transformation algorithm of Imaris XT. (C) Histogram depicting the distances of 32 clones to the surface of the spleen. Time sequences recorded in the lymph node 14 days after transplantation of Mix 5FPS Lin− HSPCs using resonant scanner were transformed into 4D movies and analyzed using XT module of Imaris software (supplemental Video 16). (D) Example of cells computationally segmented as “spots” objects and the lymph node capsule as a “surface” object. Cells expressing tdTomato, EGFP, or Cerulean were represented by magenta (M), green (G), or white (W) spots, respectively. (E) Distances of individual cells/spots to the surface were plotted according to the FP expressed (M, G, W) for comparison (the lines show the mean). (F) Cell migration trajectories (tracks) over time and displacement vectors can be analyzed. (G) Distances between individual tracks and their displacement in x,y,z directions reveal cell motility heterogeneity in depth (color coded).
To explore the potential of this methodology for imaging tissues dynamically, using a time-lapse 4D approach, we imaged popliteal lymph nodes at day 14 after transplantation. Using the resonant scanner, our imaging rate was sufficiently fast to capture highly motile cells in the LN without significant motion blur, to depths comprising 10-12 layers of cells (supplemental Video 16). These time sequences were suitable to quantitate speed, trajectories, and displacement of motile cells individually and as populations (Figure 6D-G). We further explored the possibility of collecting in multicolor channels using 2-photon microscopy alone, which could enable intravital imaging. We compared imaging of a LN from a mouse transplanted with cells coexpressing 3FPs (Cerulean, Venus, tdTomato) using the TiSa laser tuned at 860 nm alone, or simultaneously with the OPO laser tuned at 1130 nm, which allowed deeper penetration of the tissue and improved excitation of the red-shifted tdTomato FP (supplemental Videos 17 and 18). These images reveal the substantial benefit of longer wavelength excitation in achieving much lower background and better visualization of tdTomato fluorescence (supplemental Figure 6).
Discussion
We present here a powerful new methodology for clonal cell tracking combining the large diversity of color-marking by 5FP-encoding LeGO vectors with in tandem confocal and 2-photon microscopy in volumetric and dynamic imaging in live tissues. A key advantage of our method is that we have extended the use of FP-based color marking by recording confocal spectral identity in 5 “distinct” 8-bit channels. Thus, the relative ratio of Cerulean, EGFP, Venus, tdTomato, and mCherry (based on our novel observation of mCherry photoconversion to a red-shifted species) within each cell creates a quantitative “colorprint” for unbiased identification of clonal identity. Combined with 2-photon excitation of intrinsic structural features of various tissues and organs, we can track color-marked clonal cells in their native environment without the need for fixation and physical sectioning, allowing us to study the ontogeny, distribution, contacts, and competitive interactions among cells within large volumes of dense tissue samples.1,8 Our approach combines the benefits of single-cell resolved high-resolution imaging together with optical sectioning via confocal microscopy. These high-resolution images from optical sections can be used to computationally reconstruct (automatically and “on-the-fly”) complete 3D volumes of great complexity to depths of ∼ 300-μm, comprising ∼ 20-30 layers of cells, vascular, bone, and collagen structures. 3D reconstructions can be used for morphometric noninvasive quantitative analyses of biologic interest. Furthermore, we prove the feasibility of high-resolution 4D live dynamic studies by combining resonant scanning multiphoton and confocal time-lapse imaging. This allows not only static characterization of various clones but also analysis of kinetic differences among cells marked by same color. We also demonstrate video-rate 4D imaging of 3FP-labeled samples using successfully in tandem TiSa and OPO lasers for higher efficiency, less harmful, deeper penetration of live tissue, paving the way for 2-photon intravital imaging. Finally, we establish a valuable option to track cells over prolonged periods (months) overcoming current dye-based limitations.32 Our approach uses commercially available confocal and 2-photon laser microscope and automated user-interactive image analysis methods based on a commercially available software package allowing easy implementation in usual microscopy facilities.
To date, a related approach termed RGB color marking using 3FPs encoding LeGO vectors has been used for clonal tracking of hepatic regeneration.13 Although this elegant work demonstrated proof of principle for combinatorial color marking via vector transduction, the studies were limited by reliance on imaging with a charged-coupled device camera of physically cut, chemically fixed tissue sections. Generating overlapping sections to cover large areas of tissues is technically challenging and labor-intensive, as is generating serial sections. Chemical fixation of soluble FPs in tissues is also problematic because of fluorescence extraction–induced changes in color combination. Using “true colors” for displaying the large palette of color marking facilitates visual inspection. Although we rely on pseudo-colors in displaying images, which could result in similar difficult to distinguish hues, we collect quantitative 5-channel “true 5FP” fingerprints that can be used to rigorously establish clonal relationships.
We apply our marking and imaging methodology to map the clonal output of individual stem and progenitor cells during hematopoietic reconstitution of the BM, and hematopoietic and nonhematopoietic organs after transplantation. During the first several weeks after administration of HSPCs to an irradiated recipient mouse, highly polyclonal hematopoiesis can be visualized in both the BM and spleen, as demonstrated by numerous cellular clusters of distinct colors. By one month after transplantation, the diversity of clonal contributions became more limited, as short-term repopulating clones with little self-renewal potential are replaced by a much lower number of long-term repopulating stem cells. The monochromatic pattern in large volumes of marrow, despite multiple clones still being active as evidenced by imaging of the peripheral blood, suggests that each clone and its progeny remain localized and spread contiguously in the marrow space.
The majority of clones detected in animals long-term contained only one or 2FPs compared with short-term clones repopulating the BM, blood, and spleen in the first several weeks after transplantation, which showed a more diverse array of FP combinations. We found no evidence for silencing of vectors with proliferation or over time as an explanation because individual macroscopic CFU-S imaged in the spleen demonstrated that all cells in each clone, despite massive proliferation, maintained the same spectral fingerprint, and clones in secondary transplants of marrow collected from primary animals 4 months after transplantation continued to express the original FPs. It is conceivable that high expression of multiple FPs is toxic to long-term repopulating HSCs; thus, clones expressing multiple FPs do not engraft long-term. However, we think that the most likely explanation for lower color diversity over time is lower efficiency of transduction of long-term HSCs versus short-term HSCs and committed progenitors. Experiments are in progress to further investigate this possibility.
The use of a strong internal viral promoter to drive FP expression raises concerns that oligoclonality long-term resulted from clonal dominance of cells with insertions activating adjacent proto-oncogenes. However, comparative studies have demonstrated that even strong viral promoters located internally within third-generation lentiviral vectors are very unlikely to impact on hematopoietic clonal behavior and if oncogene activation were contributing, it would be more likely for clones with multiple insertions to dominate.33,34 Our findings are complementary to molecular studies tracking clonal output of HSPCs via genetic tagging; early polyclonal reconstitution followed by a more limited number of long-term repopulating clones, at least in the murine setting after total body irradiation.35,36 It is also possible in both these prior studies and in the current study that the in vitro culture required to transduce cells resulted in significant losses in long-term repopulating cell numbers or function, contributing to oligoclonality. However, molecular approaches do not provide insights into the spatial distribution of hematopoiesis or allow analysis of the interaction between clone behavior and environmental cues. Understanding these aspects of the development of hematopoietic clones in an irradiated reconstituted animal, in addition to its intrinsic interest, is of clinical relevance for chemotherapy, transplantation, and gene therapy.
The ability of HSPCs to give rise to cell types not traditionally associated with their lineage is a subject of intense interest.37-40 To date, in vivo studies evaluating the potential of stem cells have reported conflicting findings.41-48 Our approach allows clear tracking of color-marked hematopoietic clones readily identifiable in the tissues of recipient for extended periods of time, bringing a new tool for assessing potential contributions of HSPCs to other organs and tissues. The strong internal viral promoter has been demonstrated to provide high-level constitutive expression in both hematopoietic and nonhematopoietic tissues.13,49-51 In contrast to prior studies, where serial sections were used in an attempt to colocalize hematopoietic and tissue-specific markers, such as CD45 and keratin, the use of confocal microscopy and 3D reconstruction gives much less ambiguous results. Thus far, we have not observed FPs introduced via Lin− BM cell transduction expressed in skeletal muscle fibers, cardiomyocytes, hepatocytes, epidermis, or other nonhematopoietic tissues.
In conclusion, our approach is not limited to the analysis of HSPCs. Many biologic systems could benefit from the ability to resolve spatiotemporal arrangements of clonally complex cellular and structural elements via multicolor labeling and confocal and 2-photon imaging. Organ regeneration, immune responses, and tumor metastatic patterns could be interrogated, to suggest just a few potential applications.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Boris Fehse (University Medical Center Hamburg-Eppendorf, Hamburg, Germany) for providing the 5 LeGO vector plasmids; Christian A. Combs and Neal S. Young (National Heart, Lung, and Blood Institute, National Institutes of Health) for discussions, support, and encouragement throughout this study; Pradeep Dagur (flow cytometry core, National Heart, Lung, and Blood Institute, National Institutes of Health) for assistance with flow cytometric analysis; Bernd Zinselmeyer (National Institute of Neurological Disorders and Stroke, National Institutes of Health) for discussions on live 2-photon imaging; and Andre LaRochelle (National Heart, Lung, and Blood Institute, National Institutes of Health) for assistance with tail vein injections.
This work was supported by the Intramural Research Program of the National Heart, Lung, Blood Institute of the National Institutes of Health.
National Institutes of Health
Authorship
Contribution: D.M. conceived and designed the microscopy methodology, performed microscopy experiments, analyzed data, prepared figures, and wrote the manuscript; J.-Y.M. prepared lentiviral vectors, performed transductions and murine transplantation experiments, performed microscopy experiments, analyzed data, and wrote the manuscript; and C.E.D. conceived the study, led the research, analyzed the results, and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Cynthia E. Dunbar, Hematology Branch, National Heart, Lung, and Blood Institute (NHLBI)/National Institutes of Health (NIH), Bldg 10-CRC, Rm 4E-5132, 9000 Rockville Pike, Bethesda, MD 20892; e-mail: dunbarc@nhlbi.nih.gov; and Daniela Malide, Light Microscopy Core Facility, NHLBI/NIH, Bldg 10, Rm 6N309, 10 Center Dr, Bethesda, MD 20892; e-mail: dmalide@nih.gov.
References
Author notes
D.M. and J.-Y.M. contributed equally to this study.

![Figure 1. Confocal microscopy approach for imaging cells transduced with LeGO vectors encoding 5FPs. NIH3T3 cells transduced with LeGO vectors encoding single FPs were used to define imaging parameters. (A) Spectral (xyλ) imaging was used to record reference emission spectra for each FP, illustrated in normalized histograms pseudo-colored with cyan (Cerulean), green (EGFP), yellow (Venus), magenta (tdTomato), and red (mCherry). The mCherry spectrum red shifts (12 nm at peak) when excited by 594 nm (solid red line) compared with 561 nm (dashed red line) wavelength. (B) Five channels were set to image sequentially: Cerulean (458 emission [em] 468-482), EGFP (488 em 496-514), Venus (514 em 523-558), tdTomato (561 em 579-597), and mCherry (594 em 618-670). (C) Imaging of NIH3T3 cells transduced with individual FP vectors. Each FP was visible only in the cells transduced with the corresponding vector imaged in the appropriate channel, and absent from the others (no crosstalk). (D-E) Populations of NIH3T3 cells transduced independently with the 5 LeGO vectors and then mixed (Mix, D) or NIH3T3 cells transduced with all 5 vectors concurrently (Co, E), plated and imaged 4-5 days later. (D) The 5 individual FPs are shown pseudo-colored cyan (Cerulean), green (EGFP), yellow (Venus), magenta (tdTomato), and red (mCherry). (E) Clusters of progeny cells inheriting a specific color within a diverse palette of colors based on combinations of 1-5 FPs at variable intensities in each originally transduced cell. Progeny inherit the specific color, allowing tracking of lineage relationships (related images; supplemental Videos 1 and 2). Scale bar represents 100 μm (D-E).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/120/26/10.1182_blood-2012-06-440636/4/m_zh89991299140001.jpeg?Expires=1765895896&Signature=PgAgnZEDmXfYx0-Uc884UsVmPacjsybftEIvSwnZQt34TKFxgefMiJn~W4RamkUb3qTn8Ak17edE23GkPqyriG~BYUU1x0lsPJMyvfnbFLOpCHDYnDznqQcutRr0msZ0FdEgRonfsxglTlEpoVEYwZS0URQ4cbGWQ0hgPvbyQF4kxx0sWS87QMBbUwuTWlgrK6HfFyeUuGj~~BNsIlx6zOJKNclaMOTYtWiWYEHO8FKo1A-ntmBpWkNvRbjCmXWsjpGBAjmD57ZNOSgI-R2-q36prpEBldTqCNbu1Bf9usO7LGq-JHW7B1mnmn6Ee-IOFINcrknNlzt-QvDyoT1ubg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal