Abstract
Delta-like 4 (DLL4), a membrane-bound ligand belonging to the Notch signaling family, plays a fundamental role in vascular development and angiogenesis. We identified a conserved microRNA family, miR-30, which targets DLL4. Overexpression of miR-30b in endothelial cells led to increased vessel number and length in an in vitro model of sprouting angiogenesis. Microinjection of miR-30 mimics into zebrafish embryos resulted in suppression of dll4 and subsequent excessive sprouting of intersegmental vessels and reduction in dorsal aorta diameter. Use of a target protector against the miR-30 site within the dll4 3′UTR up-regulated dll4 and synergized with Vegfa signaling knockdown to inhibit angiogenesis. Furthermore, restoration of miR-30b or miR-30c expression during Kaposi sarcoma herpesvirus (KSHV) infection attenuated viral induction of DLL4. Together these results demonstrate that the highly conserved molecular targeting of DLL4 by the miR-30 family regulates angiogenesis.
Introduction
DLL4 plays a fundamental role in vascular development and angiogenesis.1,2 DLL4 haploinsufficiency results in extensive arterial defects and embryonic lethality,3 indicating that the developing vasculature is sensitive to minor alterations in DLL4 dosage. DLL4 expression is mainly restricted to the endothelium of nascent vessels, particularly the tip cells, where it maintains stalk cell identity in neighboring cells, thereby regulating vessel sprouting and branching in response to angiogenic stimuli.4 The importance of optimal DLL4 expression in physiologic angiogenesis is illustrated through its regulation of intersegmental vessel (ISV) development in zebrafish. Morpholino (MO) knockdown of dll4 in zebrafish results in an increased number of endothelial cells within the ISVs and ectopic ISV branching from the dorsal aorta (DA) because of overactivation of Vegfa signaling.5,6
DLL4 is relevant in pathologic angiogenesis and is overexpressed in human tumors, often in association with markers of inflammation, hypoxia and angiogenesis.7-9 Inhibition of DLL4 suppresses experimental tumor growth by inducing nonproductive, deregulated angiogenesis.10,11 We and others have shown that DLL4 expression is up-regulated in lymphatic endothelial cells (LECs) after infection by Kaposi sarcoma herpesvirus (KSHV),12,13 an oncogenic γ-herpesvirus that is the etiologic agent of Kaposi sarcoma (KS). KS is an angioproliferative neoplasm composed of cells of endothelial origin.14 Although accurate regulation of DLL4 levels is a hallmark of angiogenesis, the mechanisms that finely regulate DLL4 expression are not completely defined. Therefore we hypothesized that, in addition to well-known transcriptional mechanisms that affect DLL4 expression, DLL4 is regulated at the posttranscriptional level.
MicroRNAs (miRNAs) are small, noncoding RNAs that influence target gene expression through mRNA degradation and translation inhibition.15 Implicated in key cellular processes, miRNAs play a role in angiogenesis and cancer.16,17 miR-27b is the only miRNA thus far implicated in DLL4 regulation18 ; however, this miRNA also regulates sprouty homologue 2 (SPRY2) and semaphorin 6A (SEMA6A), and it is unclear whether its proposed suppression of DLL4 specifically leads to vascular defects.18,19 We previously described the miRNA signature in KSHV-infected LECs (KLECs).20 These data indicated significant down-regulation of members of the miR-30 miRNA family postinfection (PI). Encoded by 6 genes and expressed from 4 distinct transcripts across the human genome, the members of the miR-30 family share an identical seed sequence and hence have common predicted targets.21 Here we show that miR-30b and miR-30c target DLL4 in vitro and in vivo, and that the miR-30 family regulates angiogenesis.
Methods
Cell culture
LECs were purchased from Promocell and grown in endothelial growth medium MV (Promocell) supplemented with 10 ng/mL VEGF-C (R&D Systems). HUVECs were purchased from Promocell and grown in endothelial growth medium MV2 (Promocell). For both LECs and HUVECs, experiments were performed before passage 8. BCBL-1 cells, latently infected with recombinant GFP-KSHV,22 were cultured as previously described.23 293T and immortalized human fibroblast cells were grown in DMEM (Invitrogen), supplemented with 10% FBS.
KSHV production and infection of LECs
KSHV was produced and used to infect LECs as previously described.23 This procedure reproducibly resulted in 30% to 50% of LECs expressing GFP 3 days after infection.
MicroRNA mimics and inhibitors
LECs and HUVECs were seeded in 6-well plates at 5 × 104 cells per well and 293T were seeded in 12-well plates at 2.5 × 104 cells per well, 16 hours before transfection. miRIDIAN miRNA mimics and inhibitors for hsa-miR-30b, hsa-miR-30c, and the negative control No. 1 (nontargeting control; Thermo Scientific) were transfected at 100nM, unless otherwise stated. Cells were harvested for RNA or protein, used for the hanging drop assay, or transfected with luciferase reporter plasmids 48 hours after transfection.
Western blotting
LECs or HUVECs were lysed in Pierce M-PER buffer (ThermoScientific). Equal amounts of protein were resolved on a 10% polyacrylamide gel. Antibodies against DLL4 (Cell Signaling Technology), GAPDH (Monoclonal 6C5, Advanced ImmunoChemical) and α-tubulin (Monoclonal B-5-1-2, Sigma-Aldrich) were detected with HRP-conjugated secondary antibodies and were quantified using ECL or ECL Plus (GE Healthcare).
Lentivirus production and infection of LECs and HUVECs
Genomic fragments containing pre–miR-30b and pre–miR-30c-1 were cloned from LECs and were expressed using a modified pSIN-MCS lentiviral vector as described,23 subsequently referred to as pSIN_30b and pSIN_30c, respectively. The number of lentiviral copies per cell was determined by qPCR and miRNA expression was confirmed by qRT-PCR. Cells were infected in suspension. Experiments were performed 2 to 3 days after infection.
Quantitative PCR and quantitative RT-PCR
Genomic DNA for qPCR was extracted using the QIAamp DNA mini-kit (QIAGEN). The number of lentiviral copies per cell (c/c) was determined as previously described.23 Total RNA was extracted using the miRNeasy mini-kit (QIAGEN) and subjected to DNase I treatment (QIAGEN). For zebrafish work, RNA was collected from at least 10 embryos per condition or time point. Approximately 50 to 1000 ng of total RNA were used for cDNA synthesis using the SuperScript II reverse transcriptase (Invitrogen). GAPDH (housekeeping reference gene) and SELE mRNA levels were quantified by qRT-PCR using optimized forward and reverse primers at 0.3μM and SYBR Green PCR master mix (Applied Biosystems). The GAPDH primers used were as follows: forward primer 5′-GGAGTCAACGGATTTGGTCGTA-3′; reverse primer 5′-GGCAACAATATCCACTTTACCAGAGT-3′. The SELE primers used were as follows: forward primer 5′-CAGCCTCAAGATCATCAGCA-3′; reverse primer 5′-ACAGTCTTCTGGGTGGCAGT-3′. qRT-PCR quantification of DLL4, dll4, bactin1, miR-30b, and miR-30c was performed using Taqman gene expression or Taqman microRNA assays (Applied Biosystems). Quantification of pre–miR-30c-1 and pre–miR-30c-2 was performed using miRNA qRT-PCR Kit and Primer Set (GenoExplorer).
Luciferase reporter assay
The reporter plasmids (50 ng), either empty vector (pEZX-MT01) or the DLL4 3′UTR containing plasmid (pEZX-DLL4), were transfected into 293T cells, 48 hours after transfection with miRNA mimic. Cells were harvested 24 hours after transfection according to the Dual-Luciferase Reporter assay system (Promega). Luciferase activity was measured using a Fluoroskan Ascent FL luminometer (ThermoScientific). Firefly activity was normalized to internal renilla luciferase levels.
3-D spheroid in vitro angiogenesis assay
HUVECs were transfected with miRNA mimic (ThermoScientific) and after 24 hours spheroids were generated as previously described.24 One hundred spheroids were generated per condition, collected after 24 hours and embedded in matrigel basement membrane matrix (BD Bioscience). Spheroids were monitored for 120 hours and photographs were taken on an Axiovert 100 microscope (Zeiss) using an AxioCam (Zeiss) and acquired using AxioVision software (Zeiss). To analyze average sprouts per spheroid, sprouts were counted using Adobe Photoshop CS2 (n = 60). Average sprout length was measured using the segmented lines tool in ImageJ (National Institutes of Health). Five sprouts were measured per spheroid (n = 20).
Embryo manipulation and in situ hybridization procedures
Zebrafish embryos were obtained by natural spawning of adult zebrafish. Embryos were raised and maintained at 28.5°C in system water and staged as described.25 Tg(kdrl:EGFP)26 and Tg(fli1a:EGFP)27 lines were used to monitor blood vessel development. Antisense MOs (GeneTools) and miRNA mimics (Thermo Fischer Scientific) were injected into 1 to 4-cell-stage embryos. The MOs used in this work were MO1-dll4 (5 ng),6 dll4-TPmiR-30 5′-TGTAAACAATCCAGAAAAAAAGATT-3′ (10 ng unless otherwise stated), dll4-TPcontrol 5′-ATAGCACTCTATTTAACTCTTTTAA-3′ (10ng unless otherwise stated), kdr MO28 (4.5 ng), and kdrl MO28 (4.5 ng). dll4-TPmiR-30 was designed so that the 3′ end binds to the miR-30 target site within the dll4 3′UTR, whereas the 5′ region binds to the downstream flanking sequence, as per Choi et al.29 dll4-TPcontrol was designed so that it binds to another unrelated region of the dll4 3′UTR (596-620 nt), which is not predicted to contain any other miRNA binding sites, as per Choi et al.29 miRNA mimics were injected in the quantities stated. In situ hybridization was performed as described.30 RNA probes were labeled with digoxigenin (Roche) and detected using BM Purple (Roche). Images of in situ hybridizations were taken using a Nikon 1200F camera on a Nikon E1500 dissecting scope and acquired using ACT-1 software (Nikon). Fluorescent images of the Tg(kdrl:EGFP) and Tg(fli1a:EGFP) embryos were taken using an Axiocam (Zeiss) on an Axiovision Lumar V2 dissecting scope (Zeiss) and acquired using AxioVision Release 4.8.0 software. Figures were generated with Adobe Photoshop CS4.
Statistical and bioinformatics analysis
All experiments were performed in independent replicates and error bars correspond to SEM unless otherwise stated. Statistical significance (P value) was calculated with a 2-sided unpaired student t test unless otherwise stated. Processing and statistical analysis of the KLEC gene expression microarray (GEM) data (GSE17016) was performed using Bioconductor packages (affy, limma)31 for the R programming language. GEM and RNAseq data for analysis of the correlation between DLL4 and miR-30 in tumors compared with normal tissue were obtained from The Cancer Genome Atlas (www.cancergenome.nih.gov) data portal (https://tcga-data.nci.nih.gov/tcga). As data for each sample were individually processed rather than normalized as a batch and because data from different platforms and data types were analyzed, both the miRNA and gene expression data were scaled (ie, between 0 and 1) and centered (ie, approximately 0) before merging. For a full list of the sample numbers, data types, and platforms, please see supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Results
KSHV regulates expression of a miRNA family predicted to target DLL4
KSHV infection of endothelial cells, including LECs, is a tractable model to study aspects of endothelial cell biology.14,32 DLL4 is one of the most significantly up-regulated genes in KLECs.12 We therefore examined the most down-regulated cellular miRNAs in KLECs to determine whether we could identify miRNAs that regulate DLL4.20 We reanalyzed our microarray data with respect to down-regulated miRNAs and found that the miR-30 family is down-regulated in KLECs 72 hours PI (Figure 1A). The most significantly suppressed probes corresponded to miR-30b and miR-30c and the microarray data were validated by qRT-PCR (Figure 1B). The mature miR-30c miRNA detected by the microarray can be produced from 2 distinct precursor hairpins (pre–miR-30c-1 and pre-miR-30c-2) and we confirmed down-regulation of both sources in KLECs (supplemental Figure 1A).
KSHV regulates expression of the miR-30 family, which is predicted to target DLL4. (A) Heatmap representing relative changes in expression of hsa-miR-30 family members in LECs after KSHV infection. Red and yellow denote low and high expression, respectively. Four replicates of LECs and KLECs are shown. Two to 3 probes are shown for each member of the miR-30 family. Probes for hsa-miR-30b and hsa-miR-30c showed significant changes in expression (**Q < .01; ***Q < .001). Original GEM data from Lagos et al.20 (B) Down-regulation of miR-30b and miR-30c in KLECs, confirmed by qRT-PCR (means + SEM, n = 3). Expression is relative to LECs. Differences between LECs and KLECs were significant (**P < .01). (C) Complementarity between miR-30 family members and the DLL4 3′UTR. Black lines indicate canonical Watson and Crick base-pairing, gray lines indicate G:U wobbles. The predicted target site within the DLL4 3′UTR, positions 59 to 66, is shown in red; miR-30 seed region is shown in green. (D) Heatmap table displaying the correlation coefficient R between expression of DLL4 and each member of the miR-30 family in the indicated tumor types. GEM and RNAseq data were obtained from The Cancer Genome Atlas (www.cancergenome.nih.gov) data portal (www.tcga-data.nci.nih.gov), as described in “Methods.” Because of the large difference in the number of replicates for each tumor type, the significance of R was calculated for each mir-30 versus DLL4 combination. The values and fill-in color indicate the degree of negative correlation. Nonsignificant correlations are grayed out; significant correlations are marked with an asterisk (*P < .05; **P < .01). Tumor types included are breast invasive carcinoma (BRCA), colon adenocarcinoma (COAD), glioblastoma multiforme (GBM), renal clear cell carcinoma (RCC), lung adenocarcinoma (LUAD), rectum adenocarcinoma (READ), and ovarian serous cystadenocarcinoma (OV).
KSHV regulates expression of the miR-30 family, which is predicted to target DLL4. (A) Heatmap representing relative changes in expression of hsa-miR-30 family members in LECs after KSHV infection. Red and yellow denote low and high expression, respectively. Four replicates of LECs and KLECs are shown. Two to 3 probes are shown for each member of the miR-30 family. Probes for hsa-miR-30b and hsa-miR-30c showed significant changes in expression (**Q < .01; ***Q < .001). Original GEM data from Lagos et al.20 (B) Down-regulation of miR-30b and miR-30c in KLECs, confirmed by qRT-PCR (means + SEM, n = 3). Expression is relative to LECs. Differences between LECs and KLECs were significant (**P < .01). (C) Complementarity between miR-30 family members and the DLL4 3′UTR. Black lines indicate canonical Watson and Crick base-pairing, gray lines indicate G:U wobbles. The predicted target site within the DLL4 3′UTR, positions 59 to 66, is shown in red; miR-30 seed region is shown in green. (D) Heatmap table displaying the correlation coefficient R between expression of DLL4 and each member of the miR-30 family in the indicated tumor types. GEM and RNAseq data were obtained from The Cancer Genome Atlas (www.cancergenome.nih.gov) data portal (www.tcga-data.nci.nih.gov), as described in “Methods.” Because of the large difference in the number of replicates for each tumor type, the significance of R was calculated for each mir-30 versus DLL4 combination. The values and fill-in color indicate the degree of negative correlation. Nonsignificant correlations are grayed out; significant correlations are marked with an asterisk (*P < .05; **P < .01). Tumor types included are breast invasive carcinoma (BRCA), colon adenocarcinoma (COAD), glioblastoma multiforme (GBM), renal clear cell carcinoma (RCC), lung adenocarcinoma (LUAD), rectum adenocarcinoma (READ), and ovarian serous cystadenocarcinoma (OV).
We used the TargetScan prediction algorithm to identify miR-30 targets and ranked them according to total context score.33 We analyzed this list with respect to genes significantly altered in KLECs and discovered that the 3′UTR of DLL4 scored favorably (total context score −0.39). TargetScan analysis of the DLL4 3′UTR sequence indicated that the miR-30 family is the best scoring miRNA for this 3′UTR (Figure 1C).
The miR-30 target site is an 8mer which is absolutely conserved within the DLL4 3′UTR across 22 species (supplemental Figure 1B). miRNAs are known to target Notch components during tumor development,34 but a function for this cross-talk in tumorigenesis is unclear. Down-regulation of the miR-30 family is associated with enhanced tumorigenesis in breast cancer and anaplastic thyroid carcinoma.35,36 However, an association between DLL4 and miR-30 expression is not described. We analyzed mRNA and miRNA expression data from the The Cancer Genome Atlas (www.cancergenome.nih.gov), specifically studying angiogenic solid tumors. We found a significant negative correlation between 1 or more miR-30 family members and DLL4 expression in breast adenocarcinoma, ovarian serous cystadenocarcinoma, and the highly angiogenic renal clear-cell carcinoma (Figure 1D). Most of the other tumors analyzed also displayed a negative correlation, although nonsignificant (Figure 1D).
miR-30b and miR-30c regulate DLL4
To validate our target predictions for miR-30b and 30c, we measured DLL4 expression after transfecting synthetic miR-30b and miR-30c mimics into LECs. DLL4 mRNA and protein levels were significantly reduced in LECs expressing either mimic (Figure 2A-B) and this effect was dose-dependent (supplemental Figure 2A). Coexpression of miR-30b and miR-30c, at an equivalent total concentration, did not increase DLL4 repression, suggesting that there are no additive or synergistic effects between these miR-30 family members (supplemental Figure 2A). These findings correspond with our target prediction studies, which indicate only 1 miR-30 target site on the DLL4 3′UTR (Figure 1C). Expression of an unrelated mRNA (SELE) was unchanged (Figure 2A), suggesting that the suppression of DLL4 mRNA does not reflect a nonspecific effect of these mimics on global mRNA levels.37
miR-30b and miR-30c regulate DLL4.DLL4 (A-C-D) and SELE (A) mRNA expression, as measured by qRT-PCR, in LECs (A-C) or HUVECs (D) transfected with mimics (A) or infected with lentiviruses (C-D). DLL4 protein expression, as measured by Western blotting, in LECs transfected with mimics (B), infected with lentiviruses (C), or transfected with inhibitors (E). Values indicate intensity of DLL4 antibody ECL plus signal normalized to GAPDH antibody ECL signal. (A) Expression is relative to nontargeting control (NTC) mimic (means + SEM, n = 4). (B) Top panel: Representative Western blots, 20 minutes' exposure of DLL4 blot and 1-second exposure of GAPDH blot. Bottom panel: intensity is relative to NTC mimic (means + SEM, n = 3). (C) Top panel: Representative Western blots, 10 minutes' exposure of DLL4 blot and 1-second exposure of GAPDH blot. Bottom panel: expression is relative to empty vector, pSIN_MCS (means + SEM, n = 4). (D) Expression is relative to empty vector, pSIN_MCS (means + SEM, n = 3). (E) Top panel: representative Western blots, 30 minutes' exposure of DLL4 blot and 1-second exposure of GAPDH blot. Bottom panel: Intensity is relative to NTC inhibitor (means + SEM, n = 2). (F) Reporter assay indicating the response of WT or mutant (mut) DLL4 3′UTR to exogenous miR-30b and miR-30c (means + SEM, n = 3). Firefly expression was normalized to renilla expression to give the relative light units (RLU), which are shown relative to NTC mimic. MT01 is a control reporter, lacking a 3′UTR sequence but containing the firefly and renilla luciferase genes. Related statistically significant values are indicated by horizontal bars. In all panels, statistical significance denoted by *P < .05; **P < .01; ***P < .001.
miR-30b and miR-30c regulate DLL4.DLL4 (A-C-D) and SELE (A) mRNA expression, as measured by qRT-PCR, in LECs (A-C) or HUVECs (D) transfected with mimics (A) or infected with lentiviruses (C-D). DLL4 protein expression, as measured by Western blotting, in LECs transfected with mimics (B), infected with lentiviruses (C), or transfected with inhibitors (E). Values indicate intensity of DLL4 antibody ECL plus signal normalized to GAPDH antibody ECL signal. (A) Expression is relative to nontargeting control (NTC) mimic (means + SEM, n = 4). (B) Top panel: Representative Western blots, 20 minutes' exposure of DLL4 blot and 1-second exposure of GAPDH blot. Bottom panel: intensity is relative to NTC mimic (means + SEM, n = 3). (C) Top panel: Representative Western blots, 10 minutes' exposure of DLL4 blot and 1-second exposure of GAPDH blot. Bottom panel: expression is relative to empty vector, pSIN_MCS (means + SEM, n = 4). (D) Expression is relative to empty vector, pSIN_MCS (means + SEM, n = 3). (E) Top panel: representative Western blots, 30 minutes' exposure of DLL4 blot and 1-second exposure of GAPDH blot. Bottom panel: Intensity is relative to NTC inhibitor (means + SEM, n = 2). (F) Reporter assay indicating the response of WT or mutant (mut) DLL4 3′UTR to exogenous miR-30b and miR-30c (means + SEM, n = 3). Firefly expression was normalized to renilla expression to give the relative light units (RLU), which are shown relative to NTC mimic. MT01 is a control reporter, lacking a 3′UTR sequence but containing the firefly and renilla luciferase genes. Related statistically significant values are indicated by horizontal bars. In all panels, statistical significance denoted by *P < .05; **P < .01; ***P < .001.
As the highly stable mimics led to a supraphysiologic up-regulation of miR-30 of more than 1000-fold (data not shown), we also used lentiviruses to overexpress miR-30 in its pre-miRNA form. Infection of LECs with lentiviruses expressing miR-30b or miR-30c (pSIN_30b or pSIN_30c) suppressed DLL4 (Figure 2C). This was confirmed in another type of endothelial cell, HUVECs (Figure 2D). We confirmed expression of the mature miRNAs in LECs and HUVECs transduced with these viruses (supplemental Figure 2B-C) and DLL4 suppression increased with increasing viral copies per cell (supplemental Figure 2B). Conversely, transfection of hairpin inhibitors against miR-30b and miR-30c into LECs led to an increase in DLL4 protein levels (Figure 2E). Taken together, these data indicate that the miR-30 family regulates endogenous DLL4.
To confirm that these miRNAs act through the DLL4 3′UTR we used a vector with the luciferase coding sequence up-stream of the DLL4 3′UTR (DLL4_wt). We expressed this construct in the presence of miR-30b or miR-30c mimics (Figure 2F) and observed a 50% reduction in luciferase activity. The control vector maintained luciferase activity in the presence of exogenous miR-30, suggesting the changes in activity were because of the action of miR-30 on the DLL4 3′UTR. To confirm this we mutated the predicted miR-30 target site in the DLL4 3′UTR (supplemental Figure 2D) to prevent miRNA association (DLL4_mut). Luciferase activity was significantly increased to near-baseline levels (Figure 2F). These data indicate that miR-30 influences the expression of DLL4 in endothelial cells by targeting a predicted site within its 3′UTR.
miR-30 targeting of DLL4 influences endothelial cell behavior in vitro
DLL4 expression influences angiogenic pathways in endothelial cells and contributes to the angiogenic signature of KSHV-infected endothelial cells.12,38 We therefore used KSHV-infection of LECs to investigate the effect of miR-30 on DLL4 levels. (Figure 3A-B). DLL4 mRNA expression was increased 3-fold in KLECs compared with LECs and this induction was attenuated to 2-fold in KLECs expressing exogenous miR-30, suggesting that miR-30 can suppress DLL4 levels in a dynamic system where it is normally up-regulated (Figure 3A). This was also reflected at the protein level where KSHV infection induced a 5-fold increase in DLL4 that was reduced to 4-fold in pSIN_30c transduced cells (Figure 3B). These findings reveal an additional miRNA-controlled layer of DLL4 regulation in human endothelial cells and are consistent with our previous work showing that activation of the extracellular-signal regulated kinase (ERK) pathway also drives DLL4 induction in KSHV-infected endothelial cells.12
Regulation of DLL4 by miR-30b and miR-30c has relevance in pathophysiologic settings. (A) DLL4 mRNA expression in LECs or KLECs infected with miR-30b and miR-30c–expressing lentiviruses measured by qRT-PCR (means + SEM, n = 3). Expression is relative to LECs infected with empty vector, pSIN_MCS. Differences between pSIN_MCS-infected and pSIN_30b/30c-infected KLECs were significant (*P < .05). (B) DLL4 protein expression in LECs measured by Western blotting. Values indicate intensity of DLL4 antibody ECL plus signal normalized to GAPDH antibody ECL signal and relative to KSHV−/pSIN_MCS+. (C) Left panels: Representative photographs at indicated time points of HUVEC spheroids embedded in matrigel. HUVECs were transfected with mimic before being induced to form spheroids. Photographs were taken on an Axiovert 100 microscope (Zeiss) using an AxioCam (Zeiss) at 5× magnification in phase contrast. Images were acquired using AxioVision (Zeiss). Middle panels: Quantification of total sprouts per spheroid (y-axis) at indicated time points (n = 60). Box plot indicates inter-quartile range, whiskers indicate total range, black line denotes median. Sprouts were counted using Adobe Photoshop CS2. The difference between NTC-transfected and miR-30b–transfected cells was significant (***P < .001). Right panels: Quantification of average sprout length (y-axis; means + SEM, n = 20). Average sprout length was measured using the segmented lines tool in Image J (National Institutes of Health) and are displayed on the y-axis as ×100 μm. Five sprouts were measured per spheroid. The difference between NTC-transfected and miR-30b–transfected cells was significant (***P < .001).
Regulation of DLL4 by miR-30b and miR-30c has relevance in pathophysiologic settings. (A) DLL4 mRNA expression in LECs or KLECs infected with miR-30b and miR-30c–expressing lentiviruses measured by qRT-PCR (means + SEM, n = 3). Expression is relative to LECs infected with empty vector, pSIN_MCS. Differences between pSIN_MCS-infected and pSIN_30b/30c-infected KLECs were significant (*P < .05). (B) DLL4 protein expression in LECs measured by Western blotting. Values indicate intensity of DLL4 antibody ECL plus signal normalized to GAPDH antibody ECL signal and relative to KSHV−/pSIN_MCS+. (C) Left panels: Representative photographs at indicated time points of HUVEC spheroids embedded in matrigel. HUVECs were transfected with mimic before being induced to form spheroids. Photographs were taken on an Axiovert 100 microscope (Zeiss) using an AxioCam (Zeiss) at 5× magnification in phase contrast. Images were acquired using AxioVision (Zeiss). Middle panels: Quantification of total sprouts per spheroid (y-axis) at indicated time points (n = 60). Box plot indicates inter-quartile range, whiskers indicate total range, black line denotes median. Sprouts were counted using Adobe Photoshop CS2. The difference between NTC-transfected and miR-30b–transfected cells was significant (***P < .001). Right panels: Quantification of average sprout length (y-axis; means + SEM, n = 20). Average sprout length was measured using the segmented lines tool in Image J (National Institutes of Health) and are displayed on the y-axis as ×100 μm. Five sprouts were measured per spheroid. The difference between NTC-transfected and miR-30b–transfected cells was significant (***P < .001).
During pathologic and physiologic angiogenesis, endothelial cells expressing DLL4 stimulate Notch signaling in adjacent cells and are specified as “tip” cells. Tip cells localize to the apex of the developing sprout, excluding the signal-receiving “stalk” cells, which contribute to the body of the developing vessel. Suppression of Notch signaling leads to excessive sprouting and multiple vessel branches because the tip cell phenotype is not restricted.2 We used an in vitro model of sprouting angiogenesis to investigate whether DLL4 targeting by miR-30b affected normal tip-cell behavior.24 HUVECs expressing either miR-30b mimics or nontargeting control (NTC) were induced to form spheroids which were then embedded in matrigel. Those spheroids composed of miR-30b–overexpressing HUVECs displayed an increased propensity to form sprouts of greater length (Figure 3C) indicating that miR-30 overexpression promotes angiogenic sprouting. miR-30 mimics down-regulated DLL4 mRNA in this cell type (supplemental Figure 3A-C).
Exogenous expression of miR-30 induces hyperbranching of intersegmental vessels in zebrafish
The developing zebrafish vasculature is an established model of angiogenic processes. Zebrafish Dll4 regulates angiogenic sprouting of ISVs and is detectable from 8 hours postfertilization (hpf) by RT-PCR5 ; using qRT-PCR, we observed dll4 induction between 6 hpf and 12 hpf in zebrafish embryos (supplemental Figure 4A). Expression of miR-30b and miR-30c homologs is found during development and in the adult fish.39 We observed a correlation between expression of miR-30 and dll4 from 18 hpf to 30 hpf (supplemental Figure 4A) corresponding with the temporal window during which angiogenic sprouting occurs.40
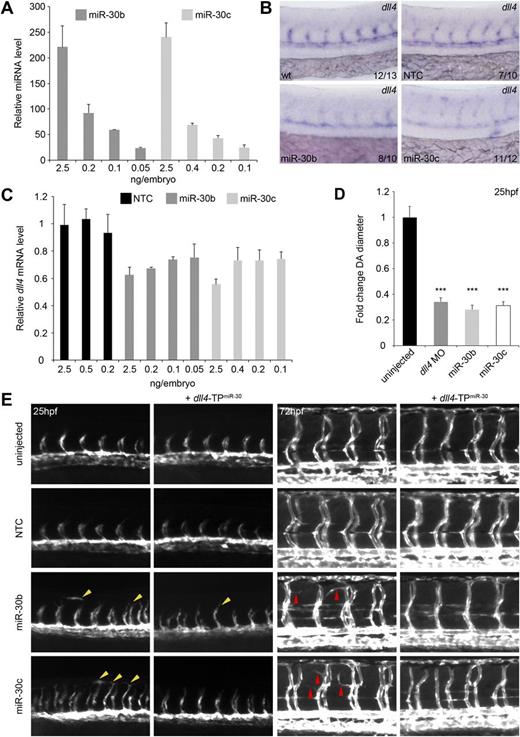
These temporally coincident changes in dll4 and miR-30 suggest a functional interaction that may contribute to tight control of Dll4 expression during vascular development. We investigated this relationship by increasing miR-30 expression through microinjection of miR-30 mimics. To reduce nonspecific effects of exogenous miR-30, microinjected mimic was titrated to levels where overall embryo morphology was normal and miR-30 expression was detectable at a physiologically relevant level (Figure 4A). We considered an up-regulation of 20- to 25-fold to be within physiologic levels, as changes of this magnitude are seen for individual miRNAs during zebrafish development.41 We confirmed the down-regulation of dll4 expression in the vasculature by in situ hybridization (Figure 4B). dll4 expression titrated with levels of miR-30 and was reduced by 20% to 30% compared with control embryos at the lowest mimic concentrations (Figure 4C). These data indicate that dll4 mRNA levels can be disrupted in vivo by exogenous expression of miR-30.
miR-30b and 30c regulate dll4 expression and angiogenic sprouting in vivo. (A) Expression of miR-30 in whole zebrafish embryos after microinjection of miR-30 mimics at the 1- to 4-cell stage measured by qRT-PCR. Expression is relative to uninjected control embryos. Values indicate the amount of injected mimic. Error bars indicate SD of qRT-PCR experiment which was performed in triplicate. n = 1 but RNA was collected from 20 to 30 embryos per condition. (B) Representative in situ hybridization showing expression of dll4 mRNA levels in the developing vasculature of WT zebrafish embryos and embryos injected with 0.05 ng (miR-30b) and 0.1 ng (miR-30c) miRNA mimics. Values indicate the number of embryos with the predominant, displayed phenotype versus the total number of embryos assayed. Images were taken using a Nikon 1200F camera on a Nikon E1500 dissecting microscope and acquired using ACT-1 software (Nikon). Images were cropped using Adobe Photoshop CS4. (C) Expression of dll4 mRNA in whole zebrafish embryos after microinjection with miR-30 mimics measured by qRT-PCR. Expression is relative to uninjected control embryos. Values indicate amount of injected mimic. Error bars indicate SD of qRT-PCR which was performed in triplicate. n = 1 but RNA was collected from 20 to 30 embryos per condition. (D) DA diameter in embryos injected with dll4 MO or miR-30 mimic relative to uninjected embryos (means + SEM, n = 3). Six DA measurements were made per embryo using Adobe Photoshop CS4. Differences between uninjected embryos and embryos injected with dll4 MO or miR-30 mimics were significant (***P < .001). (E) Trunk vasculature in uninjected Tg(kdrl:EGFP) embryos or embryos injected with the indicated miRNA mimic and/or TP. Left panels: Advanced sprouting and aberrant endothelial cell migration (yellow arrowheads) at 25 hpf. Right panels: Increased branching of the ISVs (red arrowheads) at 72 hpf. Fluorescent images were taken using an Axiocam (Zeiss) on an Axiovision Lumar V12 dissecting microscope (Zeiss) and acquired using AxioVision v4.8 software. Images were cropped and arrowheads were added using Adobe Photoshop CS4.
miR-30b and 30c regulate dll4 expression and angiogenic sprouting in vivo. (A) Expression of miR-30 in whole zebrafish embryos after microinjection of miR-30 mimics at the 1- to 4-cell stage measured by qRT-PCR. Expression is relative to uninjected control embryos. Values indicate the amount of injected mimic. Error bars indicate SD of qRT-PCR experiment which was performed in triplicate. n = 1 but RNA was collected from 20 to 30 embryos per condition. (B) Representative in situ hybridization showing expression of dll4 mRNA levels in the developing vasculature of WT zebrafish embryos and embryos injected with 0.05 ng (miR-30b) and 0.1 ng (miR-30c) miRNA mimics. Values indicate the number of embryos with the predominant, displayed phenotype versus the total number of embryos assayed. Images were taken using a Nikon 1200F camera on a Nikon E1500 dissecting microscope and acquired using ACT-1 software (Nikon). Images were cropped using Adobe Photoshop CS4. (C) Expression of dll4 mRNA in whole zebrafish embryos after microinjection with miR-30 mimics measured by qRT-PCR. Expression is relative to uninjected control embryos. Values indicate amount of injected mimic. Error bars indicate SD of qRT-PCR which was performed in triplicate. n = 1 but RNA was collected from 20 to 30 embryos per condition. (D) DA diameter in embryos injected with dll4 MO or miR-30 mimic relative to uninjected embryos (means + SEM, n = 3). Six DA measurements were made per embryo using Adobe Photoshop CS4. Differences between uninjected embryos and embryos injected with dll4 MO or miR-30 mimics were significant (***P < .001). (E) Trunk vasculature in uninjected Tg(kdrl:EGFP) embryos or embryos injected with the indicated miRNA mimic and/or TP. Left panels: Advanced sprouting and aberrant endothelial cell migration (yellow arrowheads) at 25 hpf. Right panels: Increased branching of the ISVs (red arrowheads) at 72 hpf. Fluorescent images were taken using an Axiocam (Zeiss) on an Axiovision Lumar V12 dissecting microscope (Zeiss) and acquired using AxioVision v4.8 software. Images were cropped and arrowheads were added using Adobe Photoshop CS4.
It was previously shown that dll4 silencing using MOs induces excessive ISV branching.5,6 We investigated the effect of reduced dll4 expression by miR-30 on endothelial cell behavior in Tg(kdrl:EGFP) zebrafish, using dll4 MO-injected embryos as a positive control (Figure 4E, supplemental Figure 4B). Compared with uninjected control embryos, we found that embryos expressing miR-30b or miR-30c mimics showed vessel-free, hyper-migratory endothelial cells, ISVs at a more advanced stage of sprouting and premature dorsal longitudinal anastomotic vessel (DLAV) formation at 25 hpf (Figure 4E, supplemental Figure 4B left panels yellow arrowheads). With increasing amounts of miR-30 mimic, a higher percentage of embryos displayed advanced sprouting and a greater proportion of these exhibited premature DLAV formation and hyper-migratory behavior (supplemental Figure 5A). This phenotype was also present in dll4 MO-injected embryos (supplemental Figure 4B), suggesting that miR-30 overexpression phenocopies the dll4 morphant phenotype. At 25 hpf, the diameter of the DA was significantly reduced in dll4 morphants and in miR-30–overexpressing embryos compared with WT embryos (Figure 4D). This reduction in vessel diameter concurs with previous work showing that DLL4 up-regulation in tumors correlates with vessel maturation and size.9,42 At 72 hpf, excessive ISV branching (Figure 4E, supplemental Figure 4B right panels red arrowheads) was found in embryos expressing miR-30b and 30c mimics, comparable with the branching induced by dll4 knockdown (supplemental Figure 4B red arrowheads; Leslie et al5 ). This phenotype titrated with increasing amounts of miR-30: the higher the amount of mimic injected, the greater the number of branches from the ISVs (supplemental Figure 5B).
To confirm that the miR-30 family targeted dll4 in vivo, embryos were coinjected with a target protector MO (TP) designed to bind to a region within the dll4 3′UTR containing the miR-30 target site (dll4-TPmiR-30).29 Coinjection of dll4-TPmiR-30 with miR-30b or miR-30c mimics led to a partial rescue of the miR-30–induced ISV hyperbranching (Figure 4E), with the advanced sprouting phenotype being reduced by approximately one-half in coinjected embryos (supplemental Figure 4C). These findings indicate that dll4 down-regulation is a significant contributing factor to the advanced sprouting and excessive branching phenotypes observed. These data suggest that miR-30 can regulate endothelial cell behavior in vivo and that the targeting of dll4 by miR-30 is functionally relevant during vascular development.
Increased dll4 expression synergizes with partial loss of VegfA signaling to inhibit angiogenesis
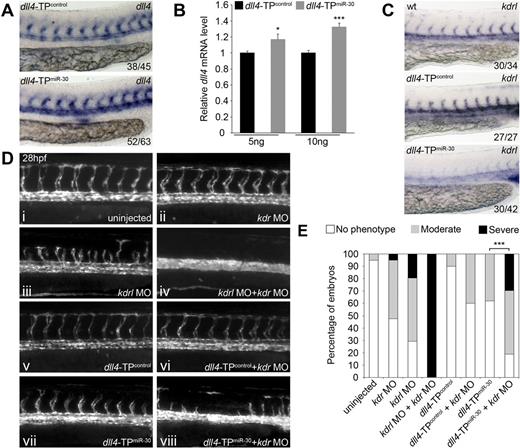
To investigate the endogenous role of miR-30 in the regulation of dll4, we injected dll4-TPmiR-30 or dll4-TPcontrol alone and examined dll4 expression at 28 hpf. Consistent up-regulation of dll4 in dll4-TPmiR-30 injected embryos was observed by in situ hybridization (Figure 5A) and qRT-PCR (Figure 5B). When Tg(fli1a:EGFP) zebrafish were injected with dll4-TPmiR-30 alone, we observed an increase in the percentage of embryos with shorter or missing ISVs at 28 hpf (Figure 5Cvii,D), suggesting that elevating dll4 levels partially blocked ISV angiogenesis.
Loss of dll4 regulation by miR-30 synergizes with partial Vegfa signaling knockdown to block angiogenesis. (A) Representative in situ hybridization showing expression of dll4 mRNA in the developing vasculature of zebrafish embryos injected with 10 ng of the indicated TP. Values indicated the number of embryos with the predominant, displayed phenotype versus the total number of embryos assayed. Images were taken using a Nikon 1200F camera on a Nikon E1500 dissecting microscope and acquired using ACT-1 software (Nikon). Images were cropped using Adobe Photoshop CS4. (B) Expression of dll4 mRNA in whole zebrafish embryos after microinjection of dll4-TPmiR-30 measured by qRT-PCR (means + SEM, n = 5). Expression is relative to embryos injected with dll4-TPcontrol. Values indicate the amount of injected TP as ng/embryo. Differences between dll4-TPcontrol and dll4-TPmiR-30 injected embryos were significant (*P < .05, ***P < .001). (C) Trunk vasculature in uninjected Tg(fli1a:EGFP) embryos or embryos injected with the indicated MO and/or TP at 28 hpf. Fluorescent images were taken using an Axiocam (Zeiss) on an Axiovision Lumar V12 dissecting microscope (Zeiss) and acquired using AxioVision v4.8 software. Images were cropped using Adobe Photoshop CS4. (D) Quantification of ISV sprouting defects after injection of the MOs and/or TPs indicated. Columns are the average of 2 independent experiments with 15 to 30 embryos counted per sample per experiment. Moderate phenotype: embryos lacking 1 ISV but with a minimum of 6 ISVs. Severe phenotype: embryos with 5 or less ISVs. To determine statistical significance, each observation of no phenotype, moderate phenotype and severe phenotype was given the value 0, 1, and 2, respectively, and then a Wilcoxon rank-sum test was performed. The difference between embryos injected with dll4-TPmiR-30 and embryos injected with dll4-TPmiR-30 + kdr MO was significant (***P < .001). Embryos were examined using an Axiovision Lumar V2 dissecting microscope (Zeiss) for quantification to be performed. (E) Representative in situ hybridization showing expression of kdrl mRNA in the developing vasculature of a WT embryo and embryos injected with 10 ng of the indicated TP. Values indicated the number of embryos with the predominant, displayed phenotype versus the total number of embryos assayed. Images were taken using a Nikon 1200F camera on a Nikon E1500 dissecting microscope and acquired using ACT-1 software (Nikon). Images were cropped using Adobe Photoshop CS4.
Loss of dll4 regulation by miR-30 synergizes with partial Vegfa signaling knockdown to block angiogenesis. (A) Representative in situ hybridization showing expression of dll4 mRNA in the developing vasculature of zebrafish embryos injected with 10 ng of the indicated TP. Values indicated the number of embryos with the predominant, displayed phenotype versus the total number of embryos assayed. Images were taken using a Nikon 1200F camera on a Nikon E1500 dissecting microscope and acquired using ACT-1 software (Nikon). Images were cropped using Adobe Photoshop CS4. (B) Expression of dll4 mRNA in whole zebrafish embryos after microinjection of dll4-TPmiR-30 measured by qRT-PCR (means + SEM, n = 5). Expression is relative to embryos injected with dll4-TPcontrol. Values indicate the amount of injected TP as ng/embryo. Differences between dll4-TPcontrol and dll4-TPmiR-30 injected embryos were significant (*P < .05, ***P < .001). (C) Trunk vasculature in uninjected Tg(fli1a:EGFP) embryos or embryos injected with the indicated MO and/or TP at 28 hpf. Fluorescent images were taken using an Axiocam (Zeiss) on an Axiovision Lumar V12 dissecting microscope (Zeiss) and acquired using AxioVision v4.8 software. Images were cropped using Adobe Photoshop CS4. (D) Quantification of ISV sprouting defects after injection of the MOs and/or TPs indicated. Columns are the average of 2 independent experiments with 15 to 30 embryos counted per sample per experiment. Moderate phenotype: embryos lacking 1 ISV but with a minimum of 6 ISVs. Severe phenotype: embryos with 5 or less ISVs. To determine statistical significance, each observation of no phenotype, moderate phenotype and severe phenotype was given the value 0, 1, and 2, respectively, and then a Wilcoxon rank-sum test was performed. The difference between embryos injected with dll4-TPmiR-30 and embryos injected with dll4-TPmiR-30 + kdr MO was significant (***P < .001). Embryos were examined using an Axiovision Lumar V2 dissecting microscope (Zeiss) for quantification to be performed. (E) Representative in situ hybridization showing expression of kdrl mRNA in the developing vasculature of a WT embryo and embryos injected with 10 ng of the indicated TP. Values indicated the number of embryos with the predominant, displayed phenotype versus the total number of embryos assayed. Images were taken using a Nikon 1200F camera on a Nikon E1500 dissecting microscope and acquired using ACT-1 software (Nikon). Images were cropped using Adobe Photoshop CS4.
Notch signaling via DLL4 negatively regulates expression of VEGFR2, the receptor through which VEGFA signals. Microarray and qRT-PCR analyzes of HUVECs transduced with empty vector and DLL4-encoding retroviruses have previously shown that VEGFR2 is down-regulated in DLL4-expressing HUVECs,38 whereas HUVECs cultured on recombinant DLL4-coated plates display reduced VEGFR2 protein levels.38 This down-regulation is known to be caused by DLL4 signaling through NOTCH as HUVECs cultured on recombinant DLL4-coated plates in the presence of the γ-secretase inhibitor DAPT did not show VEGFR2 down-regulation.38 The enzyme γ-secretase is required for the final stage of NOTCH receptor cleavage during Notch signaling.1 Zebrafish possess 2 functional orthologs of VEGFR2: kinase insert domain receptor (kdr) and kinase insert domain receptor like (kdrl).43 Knockdown of kdrl partially blocks Vegfa signaling and decreases ISV sprouting from the DA in Tg(fli1a:EGFP) embryos (Figure 5Ciii,D).28 Kdrl acts synergistically with Kdr to mediate Vegfa signaling.28 Simultaneous knockdown of kdr and kdrl causes complete loss of ISVs,28 a phenotype which we successfully recapitulated (Figure 5Civ,D). When investigating potential changes in downstream targets of Dll4 by in situ hybridization, we noted that kdrl was down-regulated on dll4-TPmiR-30 injection (Figure 5E). This suggested that the miR-30 family is indirectly regulating the levels of Vegfa signaling by negatively regulating dll4 expression which subsequently increases kdrl expression. Indeed, coinjection of dll4-TPmiR-30 with the kdr MO (Figure 5Cviii) resulted in increased loss of ISV sprouting compared with dll4-TPmiR-30 (Figure 5Cvii) or kdr MO alone (Figure 5Cii), as quantified in Figure 5D. We propose that this is caused by impairment in Vegfa signaling because of the combined down-regulation of kdr, by the kdr MO, and kdrl, by the dll4-TPmiR-30 through dll4 up-regulation. These data suggest that tight regulation of dll4 levels by the miR-30 family confers robustness to the Vegfa-mediated angiogenesis process in vivo.
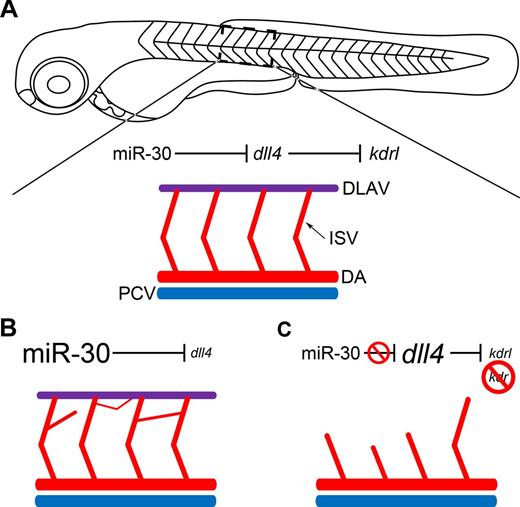
In conclusion, we show that miR-30 acts to fine tune dll4 levels during zebrafish vascular development, allowing sprouting angiogenesis to occur to the required extent (Figure 6A). When miR-30 is overexpressed, dll4 is down-regulated so tip cell fate is not restricted; this results in excessive branching from the ISVs (Figure 6B). If the interaction between miR-30 and dll4 is inhibited, the latter is up-regulated and consequently there is a reduction in kdrl expression (Figure 6C). If kdr is also suppressed, the subsequent lack of Vegfa signaling results in impaired ISV sprouting (Figure 6C).
Schematic representation of the functional consequences of DLL4 targeting by miR-30. (A) A zebrafish embryo at 72 hpf is depicted. A section of the intersegmental vessels (ISV) has been enlarged. The dorsal aorta (DA) and posterior cardinal vein (PCV) have also been depicted in red and blue, respectively. The dorsal longitudinal anastomotic vessel (DLAV) is shown in purple. (B) When miR-30 is overexpressed, dll4 is down-regulated and a deregulated network of intersegmental vessels forms as the tip cell phenotype is not restricted and excessive sprouting occurs. (C) When miR-30 regulation of dll4 is blocked using a target protector, dll4 is up-regulated, and consequently kdrl expression is reduced. When combined with kdr knockdown, the restriction of tip cell specification and inhibition of Vegfa signaling which ensues prevents normal intersegmental vessel sprouting and the DLAV does not form.
Schematic representation of the functional consequences of DLL4 targeting by miR-30. (A) A zebrafish embryo at 72 hpf is depicted. A section of the intersegmental vessels (ISV) has been enlarged. The dorsal aorta (DA) and posterior cardinal vein (PCV) have also been depicted in red and blue, respectively. The dorsal longitudinal anastomotic vessel (DLAV) is shown in purple. (B) When miR-30 is overexpressed, dll4 is down-regulated and a deregulated network of intersegmental vessels forms as the tip cell phenotype is not restricted and excessive sprouting occurs. (C) When miR-30 regulation of dll4 is blocked using a target protector, dll4 is up-regulated, and consequently kdrl expression is reduced. When combined with kdr knockdown, the restriction of tip cell specification and inhibition of Vegfa signaling which ensues prevents normal intersegmental vessel sprouting and the DLAV does not form.
Discussion
The 5 members of the miR-30 family all share the same seed sequence and are encoded by 6 genes located on human chromosomes 1, 6, and 8. This level of redundancy would suggest a critical functional role for this miRNA family. Members of the miR-30 family have been implicated in osteoblast differentiation,44 adipogenesis,45 epithelial-to-mesenchymal transition (EMT),35,46 Xenopus pronephros development,47 cellular senescence,48 myocardial matrix remodeling,49 and cancer.35,36 Genes that have been identified as targets of 1 or more miR-30 family members include snail homolog 1 (drosophila; SNAI1)46 ; LIM homeobox 1 (lhx1)47 ; B-cell lymphoma 6 (BCL6)50 ; v-myb myeloblastosis viral oncogene homolog (avian)–like 2 (MYBL2)48 ; and runt-related transcription factor 2 (RUNX2).45 Our study is the first to identify and validate DLL4 as a target of miR-30 and to demonstrate a key role for miR-30 in angiogenesis.
For sprouting angiogenesis to occur successfully a balance must be maintained between the number of tip cells, which lead the nascent sprouts, and stalk cells which will make up the endothelium of the new vessel.2 DLL4 plays a crucial role in regulating the ratio of tip cells to stalk cells. Specifically, expression of DLL4 by the tip cells stimulates Notch signaling in the adjacent cells, thereby maintaining stalk cell identity and restricting tip-cell specification.4 When DLL4 signaling or expression is inhibited, tip-cell specification is not controlled, leading to excessive sprouting from existing vessels. This can be seen during zebrafish ISV development, when knockdown of dll4 causes the formation of an aberrant network of interconnected branches.5 It is also relevant to the control of tumor vasculature as the use of blocking antibodies against DLL4 has been shown to promote angiogenesis, but the increased tumor vascularity is leaky and hence nonproductive, leading to the inhibition of tumor growth.10,11 We hypothesized that DLL4 is not only regulated at the transcriptional level but that its expression is subject to fine-tuning by posttranscriptional mechanisms, such as miRNAs, to maintain optimal levels for successful angiogenesis.
We used KSHV infection of LECs as a model with which to study DLL4 regulation, as DLL4 is one of the most significantly up-regulated genes in KLECs.12 We observed significant down-regulation of miR-30b and miR-30c in KLECs, which was of interest given that the miR-30 family was predicted to target DLL4 via a highly conserved 8mer target site within the 3′UTR. The simultaneous up-regulation of DLL4 and suppression of miR-30 in KLECs suggested a possible regulatory relationship between DLL4 and miR-30 in endothelial cells. This was further supported by our observation that DLL4 and miR-30 are negatively correlated in several tumor types that are moderately or highly angiogenic. We confirmed that DLL4 is a target of miR-30 and that miR-30 actively regulates endogenous DLL4 in human vascular and lymphatic endothelial cells in vitro and during zebrafish vascular development in vivo. We have shown, both in vitro and in vivo, that miR-30 controls DLL4 expression through a specific site within the DLL4 3′UTR. This regulation is also relevant to oncogenic virus infection, as the up-regulation of DLL4, which is normally seen in KLECs, was attenuated by the application of exogenous miR-30.
Our study has revealed a functional role for miR-30 in the regulation of angiogenesis via DLL4 targeting. Overexpression of miR-30 in endothelial cells promotes angiogenic sprouting in vitro. The introduction of exogenous miR-30 into zebrafish embryos led to aberrant migration of endothelial cells and longer sprouts at 25 hpf which developed into excessive ISV branching at 72 hpf, phenotypes also observed after dll4 knockdown. We show that the excessive vascular sprouting caused by miR-30 overexpression was by way of dll4 targeting through the use of a TP, specific to the miR-30 target site within the dll4 3′UTR. Recently it has been shown that knockdown of miR-27b causes defective vascular sprouting in developing zebrafish embryos.18 Rescue studies using knockdown of dll4 and spry2 implicated these 2 targets as the causative agents of this phenotype; however, TP studies were not used to fully delineate the respective roles of dll4 and spry2. The angiogenesis inhibitor SEMA6A has also been identified as a target of miR-27b,19 and could account for the phenotype seen on miR-27b inhibition in zebrafish embryos.
The TP also provided us with a useful tool with which to explore the endogenous role of miR-30 during zebrafish vascular development. We used it to specifically block the interaction of miR-30 with dll4 during development, while still allowing miR-30 to interact with its other targets. We have shown that inhibiting the normal control of dll4 by miR-30 elevates dll4 levels, which partially blocks ISV angiogenesis. This would suggest that during normal ISV sprouting miR-30 is actively suppressing dll4, helping to maintain tight control of dll4 expression and hence angiogenesis. However, further investigations revealed a more intricate regulatory pathway. VEGFR2 expression has been shown to be inhibited by Notch signaling via DLL4.38 When dll4 levels were increased using the TP, we observed down-regulation of kdrl, a functional ortholog of VEGFR2.43 Vegfa signaling in zebrafish is mediated through the synergistic action of Kdrl and another Vegfa receptor, Kdr.28 When injection of dll4-TPmiR-30 was combined with knockdown of kdr, ISV sprouting was moderately to severely inhibited in the majority of embryos, as seen when the expression of both kdrl and kdr is blocked using MOs.28 This work has therefore revealed that the miR-30 family indirectly regulates Vegfa signaling by controlling dll4 expression and that this regulatory axis confers robustness to Vegfa-mediated angiogenesis.
We have identified and validated DLL4 as a novel target of the miR-30 family and demonstrated the important role of these miRNAs during sprouting angiogenesis. We show that miR-30 regulates vascular development in vivo and provides robustness to Vegfa signaling by indirectly influencing kdrl expression. Our findings indicate that the manipulation of miR-30 in the setting of pathologic vascularization could represent a new therapeutic approach.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Prof A. Harris and Dr E. Bridges for advising on the 3D spheroid in vitro angiogenesis assay.
This work was funded by a United Kingdom MRC PhD Studentship (G.B.), an MRC Program Grant (C.B. and D.L.), a BHF project grant (R.M. and R.P.), an MRC Unit Grant (R.M. and R.P.), and by Cancer Research UK (S.H., V.E., and C.B.).
Authorship
Contribution: G.B. designed and performed the experiments, analyzed the data, and wrote the paper; R.M. designed and performed the zebrafish experiments and assisted in preparation of the paper; S.H. provided the bioinformatics analysis and statistical assistance; V.E. designed experiments and assisted with paper preparation; D.L. performed the miRNA microarray and assisted with paper preparation; D.G. performed some of the KSHV experiments; R.P. assisted with paper preparation; and C.B. designed the experiments, supervised the project, and wrote the paper.
Conflict-of-interest disclosure: G.B., V.E., and C.B., are joint inventors on a patent application for DLL4 targeting by the miR-30 family, which was filed at the UKPTO on 09.02.11, application No. 1102283.7. The remaining authors declare no competing financial interests.
Correspondence: Chris Boshoff, Cancer Research UK Viral Oncology Group, UCL Cancer Institute, University College London, London, WC1E 6BT, United Kingdom; e-mail: c.boshoff@ucl.ac.uk.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal