Abstract
The pathogenesis of classical Hodgkin lymphoma (cHL), the most common lymphoma in the young, is still enigmatic, largely because its Hodgkin and Reed-Sternberg (HRS) tumor cells are rare in the involved lymph node and therefore difficult to analyze. Here, by overcoming this technical challenge and performing, for the first time, a genome-wide transcriptional analysis of microdissected HRS cells compared with other B-cell lymphomas, cHL lines, and normal B-cell subsets, we show that they differ extensively from the usually studied cHL cell lines, that the lost B-cell identity of cHLs is not linked to the acquisition of a plasma cell-like gene expression program, and that Epstein-Barr virus infection of HRS cells has a minor transcriptional influence on the established cHL clone. Moreover, although cHL appears a distinct lymphoma entity overall, HRS cells of its histologic subtypes diverged in their similarity to other related lymphomas. Unexpectedly, we identified 2 molecular subgroups of cHL associated with differential strengths of the transcription factor activity of the NOTCH1, MYC, and IRF4 proto-oncogenes. Finally, HRS cells display deregulated expression of several genes potentially highly relevant to lymphoma pathogenesis, including silencing of the apoptosis-inducer BIK and of INPP5D, an inhibitor of the PI3K-driven oncogenic pathway.
Introduction
Hodgkin lymphoma (HL), one of the most frequent lymphomas, is composed of 2 entities: classical HL (cHL), accounting for ∼ 95% of cases, and the rare nodular lymphocyte predominant HL (nLPHL).1 Tumor cells are named LP cells in nLPHL and Hodgkin and Reed-Sternberg (HRS) cells in cHL. Based on differences in the histologic picture and composition of the microenvironment, cHL is subtyped into nodular sclerosis (NS), mixed cellularity (MC), lymphocyte-rich (LR), and lymphocyte-depleted (LD) cHL.1
HL is very unusual as the tumor cells often account for < 1% of the cellular population in affected lymph nodes. In cHL, the majority of cells consists of T and B cells, macrophages, and other cell types. The rarity of HRS cells and the existence of only few cHL-derived cell lines have hampered their molecular analysis. Nevertheless, studies of microdissected HRS cells demonstrated that they derive from germinal center (GC) or post-GC B cells because they carry somatically mutated immunoglobulin (Ig) V genes.2 Destructive IgV gene mutations in a fraction of cases suggest a derivation of HRS cells from GC B cells that acquired such mutations and normally would undergo apoptosis.3,4 In ∼ 40% of cHL, HRS cells are latently infected by Epstein-Barr virus (EBV), and EBV is likely capable to rescue preapoptotic GC B cells from apoptosis.5 There is intensive discussion about the relationship between cHL and B-cell non-HL (B-NHL). In particular, cHL shares with primary mediastinal B-cell lymphoma (PMBL) several genetic, immunophenotypic, and biologic characteristics, and the NS-cHL subtype often has also a similar clinical presentation.6-8 Furthermore, gene expression profiling studies showed that part of the signature distinguishing PMBL from diffuse large B-cell lymphomas (DLBCLs) is detected also in cHL cell lines.9,10 However, the histopathologic picture of these neoplasms is typically quite different. Moreover, additional biologic/diagnostic “gray zones” at the interface between cHL and other B-cell lymphomas, such as nLPHL, T cell/histiocyte-rich B-cell lymphoma (TCRBL) and DLBCL have been described.1,8,11,12
We previously performed genome-wide expression studies on cHL cell lines compared with B-NHL and normal B cells, thereby identifying genes aberrantly expressed by HRS cells and uncovering a dramatic loss of the B-cell transcriptional program in cHL.13,14 However, isolated HRS cells cultured in suspension might not faithfully portray the gene expression program of primary HRS cells in the lymph node microenvironment. Whole-tissue RNA analysis was valuable in characterizing the cHL microenvironment but not the HRS cells.15-17 We recently established an approach to perform genome-wide expression profiling of cells laser-microdissected from frozen biopsies and initially characterized the tumor cells of nLPHL and anaplastic large cell lymphoma.18,19 Here we applied this methodology to primary HRS cells to elucidate cHL pathogenesis.
Methods
Normal and neoplastic B-cell samples
The isolation of naive (N) and memory (M) B cells from peripheral blood of 5 healthy adult donors, and of CD77− centrocytes (CC), CD77+ centroblasts (CB), and plasma cells from routine tonsillectomies has been described.18 As CD77 was later shown to be unsuitable for separating centroblasts and centrocytes,20,21 we pooled CD77+ and CD77− samples as both representative of GC B cells (see supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Gene chip data for the normal B-cell samples, 5 nLPHLs, 4 TCRBLs, 11 DLBCLs (classified according to the Hans algorithm22 as 1 GCB (GC B cell-like), 8 non-GCB and 2 not evaluable), 5 follicular lymphomas (FL) and 5 Burkitt lymphomas (BL) were previously reported,18 together with those of 16 cHLs as far as it concerns only their comparison with nLPHL18 or anaplastic large cell lymphoma.19 Here we analyze the gene expression profiles of HRS cells from these 16 cHL (all sampled at disease onset, except one at relapse) and also compared them with 4 cHL cell lines (L428, L1236, KMH2, and HDLM2; the latter is T cell–derived) and with lymphoma cells from an extended panel of B-NHL, including 5 PMBLs. Approval by the Internal Review Boards in Essen and Frankfurt for these studies, conducted in accordance with the Declaration of Helsinki, was obtained.
Microdissection of lymphoma cells
Lymphoma cells were microdissected from 5-μm-thick, H&E-stained, frozen lymph node sections using the Laser Microdissection and Pressure Catapulting technique.18 HRS cells were microdissected as single cells according to their characteristic morphology. In PMBL, areas with at least 95% tumor cells were microdissected. Cells were catapulted into Purescript lysis buffer (Gentra) and pooled in groups of 1000-2000 cells.
Generation of gene expression profiles
RNA isolation, generation of cRNA by 2 rounds of in vitro transcription, cRNA fragmentation and hybridization to Affymetrix HG-U133-Plus2.0 arrays, and microarray washing and scanning were described previously.18 The gene expression dataset is available at http://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE 12453, 14879, and 40160).
Bioinformatic analysis of the microarray data
Most bioinformatics analyses were performed using GeneSpring Version 7.3.1 software (Agilent Technologies) with GC-RMA preprocessing of the raw expression data (CEL files). Further statistical analysis was done with the computing environment R.23 Additional software packages (affy, geneplotter) were taken from the Bioconductor project24 (supplemental Methods).
Immunohistochemistry
Details are given in supplemental Methods.
Results
Using microarrays interrogating ∼ 47 000 transcripts, we generated expression profiles of HRS cells isolated from lymph node biopsies of 16 cHL patients and compared them with the profiles similarly obtained from the main cHL cell lines, GC/post-GC B-cell lymphomas, and normal mature B-cell subsets.
Primary and cultured HRS cells differ extensively at the transcriptional level in their interaction with the microenvironment and proliferative attitude
As many as 2444 probe sets (1908 named genes) were highly differentially expressed (≥ 3-fold, FDR q-value < .05) in the microdissected HRS cells versus the cHL lines on supervised analysis, with 742 and 1702 probe sets, respectively, being up-regulated in the 2 cell types (not shown). Among the cell line genes, many were involved in the general cellular processes connected to cell proliferation and growth (nucleotide synthesis, DNA replication, mitosis, ribosome biogenesis; not shown), indicating a globally heightened level of these processes in cultured compared with primary HRS cells. Conversely, because we observed that the primary HRS-cell genes featured several chemokines and chemokine receptors, cell adhesion molecules, and extracellular matrix remodeling factors, we undertook an unbiased gene set enrichment analysis (GSEA) and observed that each of these processes is overall transcriptionally enriched in primary versus cultured HRS cells (supplemental Figure 1). However, the 2 cell types did not considerably differ in the overall expression of typical cHL signatures, such as the high expression of NF-κB target genes or the low/absent expression of B-cell genes (not shown). Moreover, our own primary HRS-cell signature (developed in comparison to B-NHLs; see “Supervised comparison of primary HRS cells versus B-NHLs”) readily classified all HRS cell lines as cHL while labeling the DEV nLPHL cell line as non-cHL (supplemental Table 1).
cHL represent a distinct lymphoma entity, overall closer to nLPHL than to PMBL, but with differential behavior of the cHL histologic subtypes
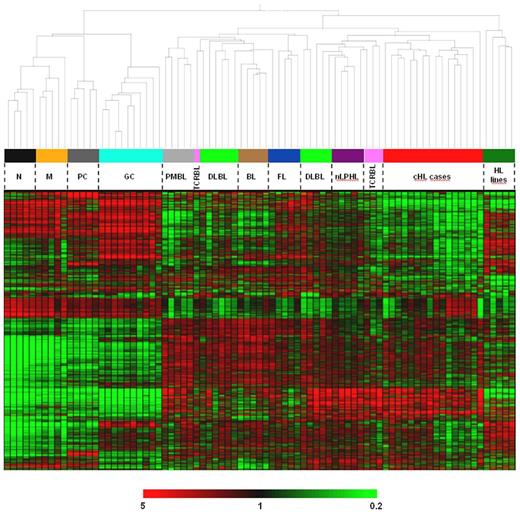
Unsupervised hierarchical clustering produced 2 main branches: one composed of all 25 normal B-cell samples (each cell type forming its own subcluster) and the other composed of all 56 neoplastic samples, with the HL lines grouping separately from the primary tumor cells (Figure 1). Among the latter, 2 sub-branches were evident: in one, cHL formed a distinct cluster (with a single “outlier,” the only cHL analyzed at relapse), separated from nLPHL (5 cases), most TCRBL (3 of 4 cases), and some DLBCL (3 of 11 cases). The observation that PMBL, BL, and cHL built the most homogeneous sub-branches without outliers, except the relapse cHL case may reflect the biologic distinctness of these lymphoma entities (ie, the known distinctness of PMBL from DLBCL,9,10 the overwhelming MYC signature in BL,25 and the uniqueness of HRS cells among B-cell lymphomas), whereas in particular for DLBCL biologic heterogeneity is well known. Overall, the clustering analysis indicates that HRS cells define a lymphoma entity characterized by a specific gene expression profile, which is distinct from other B-cell neoplasms and surprisingly globally closer to nLPHL than to PMBL, as also confirmed by the high coclustering confidence (80% on bootstrap resampling) of cHLs with nLPHLs and of PMBLs with the non-HLs in the 2 respective sub-branches of the primary tumors.
Unsupervised hierarchical clustering of normal and neoplastic B-cell samples. The dendrogram (top part) is based on the expression pattern of the 2000 probe sets (corresponding to 1535 named genes) varying the most across the dataset (and thus being the most informative); their expression pattern is depicted in the clustered heat map and color bar below (bottom part). Very similar results were obtained when using a higher or lower number of highly variant probe sets or when the clustering was restricted to the primary tumors and corresponding most variant probe sets (ie, excluding the influence of normal B cells and HL lines; not shown). In addition, the coclustering of cHL with NLPHL and of PMBL with the non-HLs was consistently observed even when restricting the analysis only to cHL, NLPHL, and PMBL or only to cHL, NLPHL, PMBL, and DLBCL (not shown). The HL line cluster is formed, from left to right, by DEV (nLPHL), HDLM2 (T-cell cHL), KMH2, L1236, and L428 (B-cell cHL). N indicates naive B cells; M, memory B cells; and PC, plasma cells.
Unsupervised hierarchical clustering of normal and neoplastic B-cell samples. The dendrogram (top part) is based on the expression pattern of the 2000 probe sets (corresponding to 1535 named genes) varying the most across the dataset (and thus being the most informative); their expression pattern is depicted in the clustered heat map and color bar below (bottom part). Very similar results were obtained when using a higher or lower number of highly variant probe sets or when the clustering was restricted to the primary tumors and corresponding most variant probe sets (ie, excluding the influence of normal B cells and HL lines; not shown). In addition, the coclustering of cHL with NLPHL and of PMBL with the non-HLs was consistently observed even when restricting the analysis only to cHL, NLPHL, and PMBL or only to cHL, NLPHL, PMBL, and DLBCL (not shown). The HL line cluster is formed, from left to right, by DEV (nLPHL), HDLM2 (T-cell cHL), KMH2, L1236, and L428 (B-cell cHL). N indicates naive B cells; M, memory B cells; and PC, plasma cells.
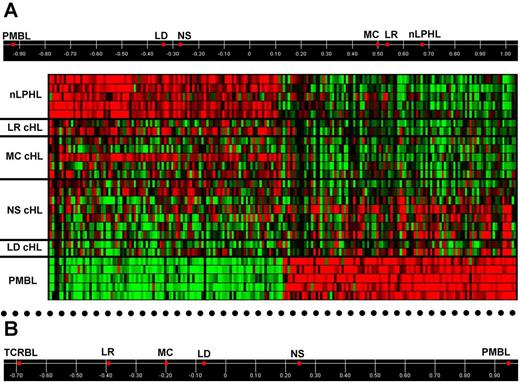
To more directly assess the relatedness of HRS cells to other lymphomas, we first confirmed by GSEA that the published transcriptional signatures distinguishing PMBL from DLBCL and shared by the cHL lines9,10 are also enriched in our PMBL versus DLBCL cells and primary HRS versus DLBCL cells (supplemental Figure 2), thus validating our data. We then derived from our profiles 168 genes highly discriminating between PMBL and DLBCL (and including several of the published ones)9,10 and used them to analyze by supervised principal component analysis (PCA) the relatedness of these lymphomas with HRS cells. For this analysis, the group of NS-cHL was kept separated from the group of the other cHL subtypes. Supplemental Figure 2 shows that NS-HRS cells map closer to PMBL than DLBCL, whereas non-NS-HRS cells are distant from PMBL and DLBCL. When the same supervised approach was applied to derive the PMBL versus nLPHL discriminating genes (Figure 2A), HRS cells of the LR and MC groups aligned very close to LP cells of nLPHL, whereas the NS and LD groups tended toward PMBL. A similar pattern was observed in the supervised PCA of PMBL versus TCRBL (Figure 2B), although LR- and MC-HRS cells appeared less close to TCRBL (Figure 2B) than to nLPHL (Figure 2A). In conclusion, HRS cells of the various histologic subtypes differ in their relatedness to other B-cell lymphomas, with LR- and MC-cHL being close to nLPHL (and, to a lesser extent, TCRBL), and NS- and LD-cHL tending toward PMBL.
Relatedness of HRS cells to PMBL and nLPHL. (A) Top: Supervised PCA using PMBL versus nLPHL discriminating genes: the top 100 probe sets significantly up-regulated (≥ 4-fold change, FDR q-value < .05) by PMBL versus nLPHL and vice-versa, which correspond to 79 and 83 annotated genes, respectively. HRS cells of the 4 cHL histologic subtypes (mean, 2-7 samples each) are aligned based on their correlation to the first principal component, which captures most of the variance (75.6%, not shown) existing between the individual PMBL (n = 5) and nLPHL (n = 5) samples in the expression of the discriminating genes and therefore has PMBL and nLPHL at its extremes. Bottom: Heat map of the discriminating genes in the individual lymphoma samples (color code identical to Figure 1); in the first 2 NS cHLs from the top, the expression pattern of both PMBL and nLPHL genes is somewhat discordant with the other 5 NS cHLs. (B) Supervised PCA of HRS cells of the 4 histologic subtypes with respect to the PMBL versus TCRBL comparison: using the top 100 probe sets significantly up-regulated (≥ 4 fold change, FDR q-value < .05) by PMBL versus TCRBL and vice-versa, which correspond to 85 and 76 named discriminating genes, respectively. Displayed is the correlation of HRS cells from the various cHL histologic subtypes with the first principal component, accounting for 77.9% (not shown) of the total variance existing between the individual PMBL (n = 5) and TCRBL (n = 4) samples in the expression of the discriminating genes.
Relatedness of HRS cells to PMBL and nLPHL. (A) Top: Supervised PCA using PMBL versus nLPHL discriminating genes: the top 100 probe sets significantly up-regulated (≥ 4-fold change, FDR q-value < .05) by PMBL versus nLPHL and vice-versa, which correspond to 79 and 83 annotated genes, respectively. HRS cells of the 4 cHL histologic subtypes (mean, 2-7 samples each) are aligned based on their correlation to the first principal component, which captures most of the variance (75.6%, not shown) existing between the individual PMBL (n = 5) and nLPHL (n = 5) samples in the expression of the discriminating genes and therefore has PMBL and nLPHL at its extremes. Bottom: Heat map of the discriminating genes in the individual lymphoma samples (color code identical to Figure 1); in the first 2 NS cHLs from the top, the expression pattern of both PMBL and nLPHL genes is somewhat discordant with the other 5 NS cHLs. (B) Supervised PCA of HRS cells of the 4 histologic subtypes with respect to the PMBL versus TCRBL comparison: using the top 100 probe sets significantly up-regulated (≥ 4 fold change, FDR q-value < .05) by PMBL versus TCRBL and vice-versa, which correspond to 85 and 76 named discriminating genes, respectively. Displayed is the correlation of HRS cells from the various cHL histologic subtypes with the first principal component, accounting for 77.9% (not shown) of the total variance existing between the individual PMBL (n = 5) and TCRBL (n = 4) samples in the expression of the discriminating genes.
EBV infection does not markedly influence the molecular profile of the established HRS cell clone
Because EBV infection probably plays an important role in cHL lymphomagenesis, we next asked if and how EBV influences the infected HRS cell at the transcriptional level. Although unsupervised clustering of the 16 cHLs produced 2 main branches (Figure 1), this segregation was not driven by the EBV status of the cases (see “Identification of 2 distinct cHL molecular subgroups, characterized by different strengths of the MYC, IRF4, and NOTCH1 activation signatures”) suggesting that in HRS cells the transcriptional changes associated with EBV are not gross. When a direct comparison between EBV+ cHLs (n = 6) and EBV− cHLs (n = 10) was performed through a more sensitive supervised approach, no genes emerged as differentially expressed in a statistically significant manner at a FDR q-value < .05, even when using a low fold-change threshold (≥ 1.5-fold). By increasing the FDR q-value to < .1 and compensating this relaxation through a greater stringency of the fold-change criterion (≥ 3-fold), only few named genes emerged as up-regulated or down-regulated (14 and 21, respectively) in EBV+ versus EBV− cHLs, including some known from the literature (supplemental Table 2; supplemental Figure 3).
HRS cells and the GC B to plasma cell transition
Regarding the relationship between HRS cells and normal B cells, we focused on the transition between GC B cells and plasma cells because HRS cells derive from GC B cells but have largely lost the pan-B and GC B-cell specific phenotype (as also plasma cells physiologically do) while acquiring expression of some plasma cell markers.26-29
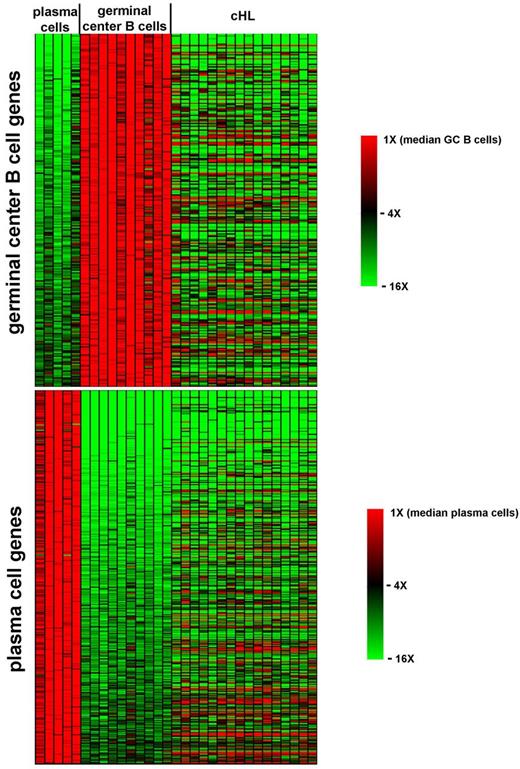
We first identified by supervised analysis of our data 625 probe sets whose expression robustly changes (≥ 5-fold change; FDR q-value < .027) during the GC B to plasma cell transition (Figure 3) and which include as internal controls (not shown) numerous known GC B-cell markers (eg, BCL6, AICDA, CD10, and IRF8) and plasma cell markers (eg, IRF4, BLIMP1, XBP1, and CD138). The expression pattern of this GC B versus plasma cell signature was then visually inspected in the profiles of primary HRS cells, which turned out to consistently down-regulate most of the GC B-cell transcriptional program but not to consistently up-regulate most of the plasma cell one (as also confirmed by PCA focusing on the 363 probe sets up-regulated by plasma cells over GC B cells; not shown). Thus, HRS cells resemble neither GC B cells nor plasma cells in their transcriptional profile, despite sharing some genetic and phenotypic features with these cell types.
HRS cells and the transition from GC B cells to plasma cells. Heat map of the expression pattern in individual plasma cell (n = 5), GC B cell (n = 10), and HRS cell (n = 16) samples of the 262 (top) and 363 (bottom) probe sets robustly up-regulated (≥ 5 fold change, FDR q-value ≤ .027) by GC B cells over plasma cells and vice-versa, respectively.
HRS cells and the transition from GC B cells to plasma cells. Heat map of the expression pattern in individual plasma cell (n = 5), GC B cell (n = 10), and HRS cell (n = 16) samples of the 262 (top) and 363 (bottom) probe sets robustly up-regulated (≥ 5 fold change, FDR q-value ≤ .027) by GC B cells over plasma cells and vice-versa, respectively.
Supervised comparison of primary HRS cells versus B-NHLs
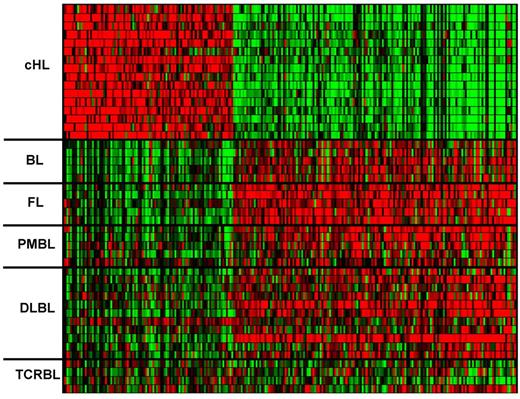
Using very stringent criteria (≥ 4-fold change, FDR q-value < .005), we identified 85 and 150 probe sets (68 and 119 named genes, respectively) up- or down-regulated in HRS cells versus B-NHL, respectively (Figure 4; supplemental Table 3). The discrimination power of these 235 cHL-specific probe sets was tested by subjecting the primary tumor dataset to class prediction (cHL vs non-cHL) using κ-nearest neighbors leave-one-out cross-validation30 (with κ = 10). Notably, such an approach, which was undertaken to validate this large-sized cHL-specific signature through an independent bioinformatic method (rather than to develop a small-sized diagnostic classifier for cHL), assigned 44 of 46 lymphomas to the correct class with high statistical significance while leaving unpredicted the remaining 2 cases (both TCRBLs; supplemental Table 1). Moreover, this signature includes several genes known7 to be up- or down-regulated in HRS cells (eg, CCL22/TARC, 40-fold up; TNFRSF8/CD30, 13-fold up; ID2, 7-fold up; AICDA 44-fold down, MS4A1/CD20 28-fold down; POU2AF1/BOB1 12-fold down; supplemental Table 3), which act as internal controls and thus give further, biologic validation to the signature.
Comparison of HRS cells with B-NHL cells. Heat map of the expression pattern in the individual primary neoplastic cell samples of the 235 probe sets discriminating between HRS and B-NHL cells (≥ 4-fold change, q ≤ .005). Color code identical to Figure 1.
Comparison of HRS cells with B-NHL cells. Heat map of the expression pattern in the individual primary neoplastic cell samples of the 235 probe sets discriminating between HRS and B-NHL cells (≥ 4-fold change, q ≤ .005). Color code identical to Figure 1.
Interestingly, the signature genes not previously known to be down-regulated in HRS cells include: (1) proapoptotic factors, such as BIK and HRK; (2) putative tumor suppressors acting in key oncogenic signaling pathways, such as RASAL1, a RAS inhibitor whose silenced expression in colon cancer correlates with lack of RAS mutations; AIM2, which can strongly inhibit NF-κB activity; and INPP5D (SHIP-1), an important inhibitor of the PI3K-AKT pathway; and (3) molecules important for chromosomal integrity, such as STAG3, a factor required for accurate chromosomal segregation and mutated in cancers with chromosomal instability (Table 1). On the other hand, novel genes up-regulated by HRS cells include RHEBL1, which induces mTOR signaling and enhances NF-κB transcriptional activity; IER3, the product of which can act as an antiapoptotic factor and ERK activator; ODZ2, encoding a transcriptional regulator overexpressed in some lymphomas because of translocation to the IgH locus; and factors involved in remodeling of and interaction with the extracellular matrix (eg, MMP12) as well as in susceptibility to chemotherapeutics effective in cHL (eg, TUBB2B and TUBB2A; Table 1).
Selected genes up- or down-regulated (≥ 4-fold; q < .005) in primary HRS cells over B-NHLs
| Gene classes and specific genes . | Fold change . | Description . |
|---|---|---|
| Apoptosis | ||
| BIK (BCL2-interacting killer) | 13.4 ↓ | BH3-only proapoptotic molecule |
| HRK (harakiri, BCL2 interacting protein ) | 4.3 ↓ | BH3-only proapoptotic molecule |
| PACAP/MGC29506 (proapoptotic caspase adaptor protein) | 10.6 ↓ | Proapoptotic molecule binding to caspase-2 and -9 |
| CYFIP2 (cytoplasmic FMR1 interacting protein 2) | 4.5 ↓ | p53-target gene inducing caspase-activation and apoptosis |
| Oncogenic signaling pathways | ||
| RASAL1 (RAS protein activator-like 1) | 10.8 ↓ | Inhibits RAS by enhancing its GTPase activity; tumor suppressor methylated in various cancers |
| DOK3 (docking protein 3) | 6.9 ↓ | Inhibits RAS by sequestering the adaptor GRB2 away from SOS (a guanine-exchange factor of RAS) |
| BTG2 (BTG family, member 2) | 4.6 ↓ | p53 target inhibiting cell cycle progression and suppressing RAS-induced transformation |
| INPP5D (inositol polyphosphate-5-phosphatase) | 4.2 ↓ | Inhibits AKT by dephosphorylating PI3P; tumor suppressor in hematologic neoplasms |
| RHEBL1 (Ras homolog enriched in brain-like 1) | 4.1 ↑ | Activates mTOR signaling and enhances NF-κB transcriptional activity |
| NLRP4 (NLR family, pyrin domain containing 4) | 9.5 ↓ | Suppresses NF-κB induction by a variety of stimuli |
| AIM2 (absent in melanoma 2) | 4 ↓ | Tumor suppressor and inhibitor of NF-κB induction |
| Interaction with the extracellular matrix | ||
| MMP12 (matrix metallopeptidase 12) | 17 ↑ | Important for tissue remodeling in various inflammatory and neoplastic disorders |
| LAYN (layilin) | 12.8 ↑ | Cell surface receptor of hyaluronan |
| CTTN (cortactin) | 5.8 ↑ | Crucial component of invadopodia in cancer cells |
| Others | ||
| ODZ2 (odz, odd Oz/ten-m homolog 2) | 33.2 ↑ | Transcriptional regulator translocated to the IgH locus in MALT lymphomas |
| IER3 (immediate early response 3) | 6 ↑ | Can potentiate ERK activation, and acquire an antiapoptotic function on phosphorylation by ERK |
| STAG3 (stromal antigen 3) | 6.2 ↓ | Component of the cohesin complex important for chromosomal stability |
| CENPV (centromere protein V) | 5.4 ↓ | Required for centromere organization, chromosome alignment, and cytokinesis |
| TUBB2B (tubulin, β 2B) | 15.3 ↑ | Tubulin heterodimers containing the tubulin β-2 isoform are the most sensitive to vinblastin |
| TUBB2A (tubulin, β 2A) | 5.2 ↑ | Tubulin heterodimers containing the tubulin β-2 isoform are the most sensitive to vinblastin |
| DNASE1L3 (deoxyribonuclease I-like 3) | 4.2 ↑ | Endonuclease sensitizing cancer cells to the topoisomerase II inhibitor etoposide |
| Gene classes and specific genes . | Fold change . | Description . |
|---|---|---|
| Apoptosis | ||
| BIK (BCL2-interacting killer) | 13.4 ↓ | BH3-only proapoptotic molecule |
| HRK (harakiri, BCL2 interacting protein ) | 4.3 ↓ | BH3-only proapoptotic molecule |
| PACAP/MGC29506 (proapoptotic caspase adaptor protein) | 10.6 ↓ | Proapoptotic molecule binding to caspase-2 and -9 |
| CYFIP2 (cytoplasmic FMR1 interacting protein 2) | 4.5 ↓ | p53-target gene inducing caspase-activation and apoptosis |
| Oncogenic signaling pathways | ||
| RASAL1 (RAS protein activator-like 1) | 10.8 ↓ | Inhibits RAS by enhancing its GTPase activity; tumor suppressor methylated in various cancers |
| DOK3 (docking protein 3) | 6.9 ↓ | Inhibits RAS by sequestering the adaptor GRB2 away from SOS (a guanine-exchange factor of RAS) |
| BTG2 (BTG family, member 2) | 4.6 ↓ | p53 target inhibiting cell cycle progression and suppressing RAS-induced transformation |
| INPP5D (inositol polyphosphate-5-phosphatase) | 4.2 ↓ | Inhibits AKT by dephosphorylating PI3P; tumor suppressor in hematologic neoplasms |
| RHEBL1 (Ras homolog enriched in brain-like 1) | 4.1 ↑ | Activates mTOR signaling and enhances NF-κB transcriptional activity |
| NLRP4 (NLR family, pyrin domain containing 4) | 9.5 ↓ | Suppresses NF-κB induction by a variety of stimuli |
| AIM2 (absent in melanoma 2) | 4 ↓ | Tumor suppressor and inhibitor of NF-κB induction |
| Interaction with the extracellular matrix | ||
| MMP12 (matrix metallopeptidase 12) | 17 ↑ | Important for tissue remodeling in various inflammatory and neoplastic disorders |
| LAYN (layilin) | 12.8 ↑ | Cell surface receptor of hyaluronan |
| CTTN (cortactin) | 5.8 ↑ | Crucial component of invadopodia in cancer cells |
| Others | ||
| ODZ2 (odz, odd Oz/ten-m homolog 2) | 33.2 ↑ | Transcriptional regulator translocated to the IgH locus in MALT lymphomas |
| IER3 (immediate early response 3) | 6 ↑ | Can potentiate ERK activation, and acquire an antiapoptotic function on phosphorylation by ERK |
| STAG3 (stromal antigen 3) | 6.2 ↓ | Component of the cohesin complex important for chromosomal stability |
| CENPV (centromere protein V) | 5.4 ↓ | Required for centromere organization, chromosome alignment, and cytokinesis |
| TUBB2B (tubulin, β 2B) | 15.3 ↑ | Tubulin heterodimers containing the tubulin β-2 isoform are the most sensitive to vinblastin |
| TUBB2A (tubulin, β 2A) | 5.2 ↑ | Tubulin heterodimers containing the tubulin β-2 isoform are the most sensitive to vinblastin |
| DNASE1L3 (deoxyribonuclease I-like 3) | 4.2 ↑ | Endonuclease sensitizing cancer cells to the topoisomerase II inhibitor etoposide |
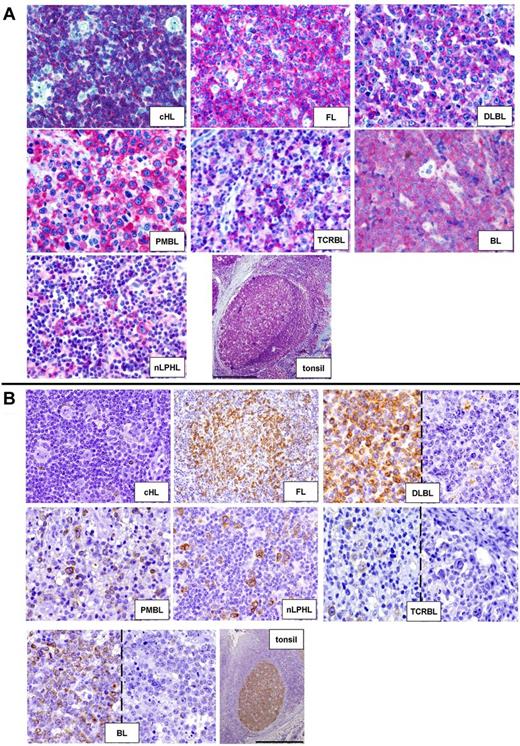
Using immunohistochemistry on paraffin-embedded biopsies (Figure 5), we validated lower overall expression of BIK and INPP5D in HRS cells versus B-NHLs (supplemental Table 4). In particular, whereas all 10 cHLs stained for INPP5D had ≤ 25% positive HRS cells, 35 of 35 and 22 of 35 B-NHLs had > 25% and > 75% positive neoplastic cells, respectively (Fisher exact test, P < .001). Similarly, 10 of 11 (91%) cHLs showed no or very rare BIK-positive neoplastic cells as opposed to only 30% (11 of 36) of B-NHLs (P < .001), and no cHL case had > 25% of positive neoplastic cells as opposed to 47% (17 of 36) B-NHLs (P < .004). When we extended such immunohistochemical survey to nLPHL (originally not included in the microarray-based comparison), we observed a higher BIK and INPP5D expression in LP versus HRS cells (Figure 5; supplemental Table 4).
Validation at the protein level of the gene expression data through immunohistochemistry. (A) HRS cells of cHL lack expression of INPP5D/SHIP-1 as opposed to neoplastic cells of the other lymphomas (40× objective) and to non-neoplastic tonsilar follicular B cells (4× objective). Bar represents 500 μm. (B) The BIK protein is absent in HRS cells of cHL, while being mostly expressed by the neoplastic cells of the other lymphomas (for DLBCL, TCRBL, and BL also examples of negative cases are shown; 40× objective). In reactive tonsils, BIK is strongly and selectively expressed by GC B cells (4× objective). Bar represents 500 μm.
Validation at the protein level of the gene expression data through immunohistochemistry. (A) HRS cells of cHL lack expression of INPP5D/SHIP-1 as opposed to neoplastic cells of the other lymphomas (40× objective) and to non-neoplastic tonsilar follicular B cells (4× objective). Bar represents 500 μm. (B) The BIK protein is absent in HRS cells of cHL, while being mostly expressed by the neoplastic cells of the other lymphomas (for DLBCL, TCRBL, and BL also examples of negative cases are shown; 40× objective). In reactive tonsils, BIK is strongly and selectively expressed by GC B cells (4× objective). Bar represents 500 μm.
Identification of 2 distinct cHL molecular subgroups, characterized by different strengths of the MYC, IRF4, and NOTCH1 activation signatures
Unsupervised hierarchical clustering of all samples revealed a subgrouping of the 16 cHL into 2 main branches (Figure 1). This was also seen when restricting the GC-RMA microarray normalization and the clustering to the cHL samples to avoid any influences of the other samples, and was observed irrespective of whether we used the 1000 (not shown), 2000 (supplemental Figure 4), or 3000 (not shown) probe sets varying the most across the 16 cHLs to perform the clustering. Further confidence about the stability of such subgrouping came from the high coclustering rate (≥ 95%) of the cHL samples belonging to each branch on multiple (n = 1000) bootstrap resampling. Importantly, this subclustering did not reflect the histologic subtype or EBV status of the cHL cases (supplemental Figure 4), nor was it detectably influenced by technical issues (eg, differences in scaling factors, present calls or 3′/M GAPDH ratios between microarrays of one versus the other branch; not shown). Supervised analysis for genes up-regulated in either of the groups revealed a surprisingly large number of > 400 at least 3-fold differentially expressed genes (FDR q-value ≤ .05; supplemental Table 5).
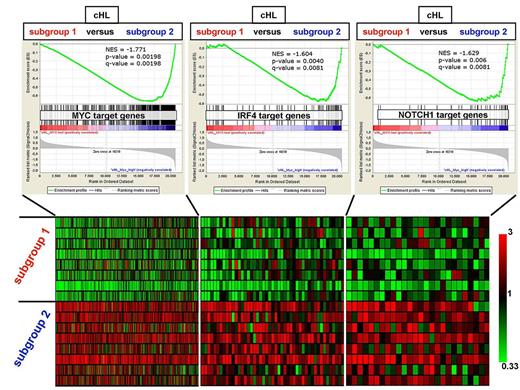
Prompted by the high mRNA expression of the MYC transcription factor we observed in some samples belonging to one branch (not shown), we used GSEA to interrogate the 2 cHL subgroups with respect to a set of 469 MYC target genes (intersection of 7054 MYC genomic binding sites and 828 genes significantly down regulated on MYC silencing) in BL cells.31 Interestingly, we observed a strong enrichment of the MYC target gene signature in one cHL subgroup versus the other, suggesting a differential transcriptional activity of MYC (Figure 6). Because IRF4 is expressed in cHL and in activated GC B-cell centrocytes in response to CD40-mediated NF-κB activity,32-34 and because myeloma cells are addicted to an IRF4 transcriptional network, which involves transactivation of IRF4 by MYC and vice-versa and which aberrantly fuses the transcriptional programs of normal plasma cells and activated B cells,35 we tested by GSEA whether the 76 IRF4 target genes in myeloma cells that are also up-regulated by normal activated B cells35 were enriched in either cHL subgroup. Interestingly, the “MYC-high” cHL cluster also displayed a heightened overall expression of this IRF4 signature (Figure 6). Furthermore, because NOTCH1 is expressed in cHL36,37 and because in T-cell acute lymphoblastic leukemia (T-ALL) cells MYC is known to be transactivated by active NOTCH1 and to cooperate with the latter in a transcriptional network promoting leukemic cell growth and cycle progression,38 we turned to the GSEA of the 44 NOTCH1 target genes in T-ALL cells.38 Notably, we observed a significant overall enrichment also of this NOTCH1 signature in the “MYC-high/IRF4-high” cHL subgroup (Figure 6). Moreover, when focusing on the leading-edge genes of the 3 signatures (ie, those genes most contributing to the differential enrichment of the respective whole signature in the 2 cHL molecular subgroups), we observed a striking proportion of IRF4 leading-edge target genes (10 of 42, 23.8%) and of NOTCH1 leading-edge target genes (13 of 27, 48.1%) overlapping with the MYC leading-edge target genes (P < .0001 by Fisher exact test).
GSEA of MYC, NOTCH1, and IRF4 target gene sets in the 2 molecular subgroups of cHL identified by unsupervised hierarchical clustering. Top: The overall expression of the target gene sets of the MYC, IRF4, and NOTCH1 transcription factors (composing 469, 76, and 44 genes, respectively) was found by GSEA to be significantly up-regulated in the profiles of microdissected HRS cells belonging to one cHL subgroup (subgroup 2; n = 8 cases) versus the other cHL subgroup (subgroup 1; n = 8 cases) emerging on unsupervised hierarchical clustering (Figure 1; supplemental Figure 4). The 44 NOTCH1 target genes in T-ALL cells are those genes whose promoters are bound by NOTCH1 and are down-regulated on pharmacologic NOTCH1 inhibition in T-ALL lines with mutated NOTCH1.38 Bottom: Heat map of the expression pattern in the individual primary HRS cell samples of the leading-edge genes (ie, those genes most contributing to the enrichment of each gene set: 264 MYC targets, 42 IRF4 targets, and 27 NOTCH1 targets) whose probe sets show ≥ 1.5-fold mean up-regulation in cHL subgroup 2. The color code of the heat map is shown by the bar on the right.
GSEA of MYC, NOTCH1, and IRF4 target gene sets in the 2 molecular subgroups of cHL identified by unsupervised hierarchical clustering. Top: The overall expression of the target gene sets of the MYC, IRF4, and NOTCH1 transcription factors (composing 469, 76, and 44 genes, respectively) was found by GSEA to be significantly up-regulated in the profiles of microdissected HRS cells belonging to one cHL subgroup (subgroup 2; n = 8 cases) versus the other cHL subgroup (subgroup 1; n = 8 cases) emerging on unsupervised hierarchical clustering (Figure 1; supplemental Figure 4). The 44 NOTCH1 target genes in T-ALL cells are those genes whose promoters are bound by NOTCH1 and are down-regulated on pharmacologic NOTCH1 inhibition in T-ALL lines with mutated NOTCH1.38 Bottom: Heat map of the expression pattern in the individual primary HRS cell samples of the leading-edge genes (ie, those genes most contributing to the enrichment of each gene set: 264 MYC targets, 42 IRF4 targets, and 27 NOTCH1 targets) whose probe sets show ≥ 1.5-fold mean up-regulation in cHL subgroup 2. The color code of the heat map is shown by the bar on the right.
Finally, as a confirmatory control, we tested by GSEA the signatures of 5 major, classical oncogenes (E2F, RAS, β-catenin, SRC, and MYC) developed in a seminal paper on oncogenic pathway signatures in human cancers39 and observed that none of them was differentially expressed in the 2 cHL groups in a statistically significant manner (nominal P < .05), except up-regulation of the MYC signature in the MYC-IRF4-NOTCH1-high cHL group (not shown). This on the one hand further supports through another independently developed MYC signature the likely different MYC activity in the 2 cHL groups, and on the other hand appears to exclude a differential activity of the other 4 classical oncogenes.
Discussion
Profiling primary HRS cells in vivo from lymph nodes taken at disease onset allowed us to compare them with cHL cell lines, a widely used (and the only available) experimental model of cHL.7 Indeed, primary and cultured HRS cells expressed similar overall levels of hallmark cHL gene signatures (eg, strong NF-κB activity and loss of B-cell identity). Moreover, the signature distinguishing primary HRS cells from B-NHLs (Figure 2; supplemental Table 1) readily identified all HRS cell lines as cHL. However, we also observed vast transcriptional differences (affecting almost 2000 named genes) between primary and cultivated HRS cells. Although we cannot exclude some influence of the different methods used for HRS cell isolation (microdissection vs sorting), other aspects are probably more important, in particular the intimate crosstalk existing in the lymph node microenvironment between the few HRS cells and the rich cellular infiltrate.7 The loss of this microenvironmental influence on the gene expression of cultured HRS cells is indeed substantiated by the transcriptional enrichment in primary HRS cells of microenvironment-related processes, such as chemotaxis, cell adhesion, and extracellular matrix remodeling, and is the result not only of the generic change from in vivo to in vitro growth but also the derivation of the available cHL lines from body fluids (eg, peripheral blood) of end-stage therapy-refractory patients, where HRS cells adapted to the growth in suspension already in vivo. This probably explains also the need to up-regulate a number of proliferation signature genes to sustain the heightened and cell-autonomous growth behavior we noted in cultured HRS cells. This concept, although applying to lymphomas in general, appears paramount in cHL, where the crosstalk with the microenvironment is most pronounced (and the attempts to establish cell lines most difficult7 ) and underscores the importance of studying lymph node HRS cells to better capture the pathogenesis of cHL. In summary, although HL cell lines have been instrumental in identifying genetic lesions and deregulated signaling pathways in HRS cells, their dramatic differences in numerous pathophysiologic features to primary HRS cells should also be kept in mind.
It should be noted that the analysis of both the HL cell lines and the primary HRS cells was focused on the bulk populations of tumor cells. There is indication from HL cell line studies that the mononuclear Hodgkin cells are the main proliferating tumor cells, whereas the Reed-Sternberg cells have little further proliferative capacity.40 There might even exist a small subset among the Hodgkin cells that fulfill some criteria of cancer stem cells, although this is controversial.41 Although it would be interesting to study such putative cells in future studies, as an initial analysis, it is essential to analyze the bulk population of the tumor cells to understand the pathophysiologic features representative of the whole lymphoma clone.
The present work also confirms in primary HRS cells (not shown) the global silencing of the B-cell program we previously uncovered in cHL cell lines.14 Because also plasma cells down-regulate many B-cell markers and HRS cells express some plasma cell transcription factors (IRF4, and inconstantly BLIMP1 and XBP1), there is a controversial discussion whether HRS cells acquired a (partial) post-GC plasma cell phenotype,26-29 perhaps pointing to a post-GC B-cell origin of HRS cells. However, we show here that HRS cells not only lack expression of most GC B-cell genes but also do not consistently express the plasma cell program (Figure 3), arguing against a plasmacytic differentiation of HRS cells and further indicating that their lost B-cell identity probably reflects a transformation-induced, aberrant coexpression in these cells of master regulators of different hematopoietic cell types.2
As infection of primary B cells by EBV causes a dramatic change in B-cell phenotype, we studied the transcriptional influence of EBV on infected HRS cells, in which the key viral genes being expressed are LMP1 and LMP2a. The fact that few cellular genes appear EBV-associated (supplemental Figure 3; supplemental Table 2) shows a relatively mild imprint left by EBV on the transcriptome of the established HRS cell clone. However, this does not exclude a major contribution of EBV in cHL pathogenesis, especially in its early steps, because the same transcriptional changes driven by EBV in infected HRS cells (eg, LMP1-induced NF-κB activity) might be reached by other mechanisms in EBV-negative HRS cells (eg, genetic inactivation of the NF-κB inhibitor TNFAIP3, which is largely restricted to EBV-negative cHL).42 Notably, a recent supervised comparison of EBV+ versus EBV− cHL cases based on whole tissue transcriptome profiling identified differentially expressed genes mainly reflecting T cell and antiviral signatures.43 Hence, EBV appears to influence the gene expression in the HL microenvironment, even though the specific impact on the infected HRS cells themselves is minor.
The current study offered us the unique opportunity to explore on a global level the transcriptional relatedness of HRS cells from the various cHL histologic subtypes to other large B-cell lymphomas, although conclusions regarding LR- and LD-cHL have to be considered as preliminary, as only 2 cases could be evaluated for each of these rare cHL subtypes (Figure 2). NS-cHL appeared more similar to PMBL cells than MC- and LR-cHL did, substantiating on a genome-wide molecular view the clinico-pathologic link between NS-cHL and PMBL.8 This is notable because none of our NS cHLs was microdissected from a mediastinal mass, and it might be that the transcriptional similarity between NS-cHL and PMBL is actually greater than we observed. Because also LD-cHL cases appeared to resemble PMBL more than MC- and LR-cHL, they could contribute together with NS-cHL to the cHL side of the interface with PMBL represented by the so-called mediastinal “gray zone” lymphomas,11,12 although this issue can be fully addressed only by also including in a comparative analysis such lymphomas (not investigated in this study). Conversely, LR-cHL appeared to be very close to nLPHL, suggesting that it has features intermediate between cHL and nLPHL not only in the microenvironment but also in the tumor cells themselves, as already appreciated regarding their morphology (smaller nuclei, less atypia) and phenotype (less frequent down-regulation of B-cell markers).44,45 It is also intriguing that the 2 cHL subtypes closest to nLPHL (ie, LR- and MC-cHL) are also the closest to TCRBL, a variant of DLBCL into which nLPHL can transform and whose reactive background (rich in T cells and histiocytes) and neoplastic cell population (sometimes cytologically indistinguishable from HRS cells and sometimes expressing CD30) can make TCRBL mimick MC-cHL.8,46 Finally, it is worth noting that, although the supervised PCA analyses shown in Figure 2 enabled us to detect some interesting transcriptional differences among HRS cells of the various cHL histologic subtypes, such differences are certainly less dramatic than the histologic differences shown by the microenvironment in these subtypes, thus pointing to genetic/epigenetic host factors as additional reasons probably modulating the immune/inflammatory response to HRS cells and producing the 4 main patterns of such response represented by the cHL histologic subtypes.
In our search for genes explaining the unsupervised clustering of cHL in 2 main branches, it was surprising to uncover that the target gene signatures of MYC, IRF4, and NOTCH1 in B-cell lymphoma, multiple myeloma, and T-ALL cells, respectively, were found to be differentially expressed between the 2 cHL groups (Figure 6; supplemental Figure 4). The expression of MYC, NOTCH1, and IRF4 by HRS cells is known and seems a general feature of all cHL histologic subtypes.32,33,37,47 NOTCH1, a T-cell transcription factor aberrantly expressed and active in HRS cells, contributes to suppressing their B-cell phenotype.36 IRF4 is crucial for plasma cell differentiation and also contributes to the down-regulation of mature B-cell genes.48 MYC is a key regulator of cell growth and proliferation.49 Intriguingly, the analysis of the leading-edge target genes of the MYC, NOTCH1, and IRF4 signatures appeared to support the concept that the same cooperation between the MYC transcriptional network on one side and those of IRF4 and NOTCH1 on the other side, which exists in myeloma cells and T-ALL cells, respectively, might be operational also in HRS cells. Immunohistochemistry confirmed the expression of MYC, IRF4, and NOTCH1 in primary HRS cells of our cHL cases but did not reveal a grossly higher protein expression in the group of cHL cases with the stronger MYC/IRF4/NOTCH1 signatures (not shown). Although immunohistochemistry is not suited to quantitatively detect subtle differences in protein expression, the differential activity of these transcription factors in the 2 cHL groups is probably mediated by other means, perhaps involving different cofactors or epigenetic changes influencing accessibility to the target genes. In addition, the association between differential expression of the MYC/IRF4/NOTCH1 signatures and the separate clustering of the 2 cHL molecular subgroups do not imply that the former is the (only) cause of the latter. There is no indication that genomic gains of the 3 genes are responsible for their increased activity in a subset of cHL, as no such alterations were found for NOTCH1 and IRF4, and only rarely for MYC.50 Thus, the cause and consequences of their differential activity in the 2 cHL molecular groups remain presently unclear but warrant clarification in future studies, considering that these factors are involved in the pathogenesis of various lymphoid malignancies.33,37
A large number of genes (187) were consistently differentially expressed (≥ 4-fold change; Table 1; supplemental Table 3) in HRS cells versus the main (post-)GC B-NHL (BL, FL, DLBCL, PMBL, and TCRBL). Immunohistochemical validation focused on BIK and INPP5D because of their interesting biologic function (BIK is a proapoptotic factor,51 and INPP5D is a phosphatase that negatively regulates PI3K-AKT signaling, to which HRS cells are dependent on for their survival and proliferation52 ) and the availability of antibodies working in routine paraffin-embedded biopsies. We confirmed the statistically significant absent or reduced expression of both genes in HRS cells compared with the B-NHL category as a whole, and we extended this also to each specific B-NHL entity, as well as to nLPHL and normal GC B cells (Figure 5; supplemental Table 3). This confirms and extends the reliability of our methodology already documented by previous studies on nLPHL and anaplastic large cell lymphoma, which used the same techniques for expression profiling and validated each of the 16 markers tested by immunohistochemistry.18,19
Because INPP5D expression is relatively restricted to hematopoietic cells53 and because HRS cells express a phosphorylated, inactive form of the ubiquitously expressed PI3K-inhibitor PTEN,52 INPP5D down-regulation might be a means to reach an undisturbed activation of the oncogenic PI3K-AKT pathway triggered in these cells by receptor tyrosine kinases, CD30, CD40, and RANKL.52,54,55 Silencing of INPP5D might possibly be mediated by mir155, known to be overexpressed in HRS cells and to cause INPP5D mRNA degradation.56,57
Among various proapoptotic factors down-regulated in HRS cells, the BH3-only molecules HRK and BIK may be interesting as they preferentially inhibit antiapoptotic BCLXL, BCLW, and BCL2A1 rather than BCL2 and MCL1.51 Intriguingly, HRS cells express more frequently BCLXL than BCL2 and MCL158,59 (BCLW and BCL2A1 were not analyzed) and might therefore need to down-regulate HRK and BIK expression to efficiently allow apoptosis inhibition through BCLXL.
In conclusion, the present study advances our understanding of HRS cell pathobiology in several important aspects. We show that cHL cell lines, although retaining key features of primary HRS cells, have undergone major gene expression changes to survive in suspension, which must be taken into consideration when studying these cell lines. We also demonstrate that the lost B-cell identity of HRS cells is not the result of the acquisition of a transcriptional plasma cell signature. The recognition that EBV infection has little specific influence on the gene expression pattern of HRS cells was an unexpected finding, as was the observation that 2 major subgroups of cHL emerged that were not defined by the histologic subtype or EBV status but were associated with a differential signature strength of the proto-oncogenes MYC, IFR4, and NOTCH1. We also clarified the transcriptional relatedness between HRS cells and the other (post-)GC B-cell lymphomas, pointing to cHL as a lymphoma entity that is overall distinct, but whose tumor cells display a differential similarity with the other lymphomas depending on the cHL histologic subtype. We finally defined a large number of HRS cell-specific genes significantly up-or down-regulated compared with the main (post-)GC B-cell lymphomas. The function of many of these genes appears highly relevant to HRS cell biology. These represent a rich source of candidate genes for future studies on the pathomechanisms of cHL and may also be useful to establish novel diagnostic markers and to propose novel therapeutic targets.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Andreas Bräuninger for helpful discussions and support and Ralf Lieberz and Nicole Diekert for excellent technical assistance.
This work was supported by Deutsche Forschungsgemeinschaft (KU1315/5-2 and 7-1). E.T. was supported by the German José Carreras Leukemia Foundation (fellowship F 05/01) and Ms Livia Benedetti (fellowship). B.F. was supported by Associazione Italiana Ricerca sul Cancro (grant MCO 10007).
Authorship
Contribution: E.T. designed and performed research, collected, analyzed, and interpreted data, performed statistical analyses, and wrote the manuscript; C.D. performed statistical analyses and analyzed and interpreted data; V.B. performed research and collected, analyzed, and interpreted data; C.J.M.v.N., W.K., G.M., B.F., and M.-L.H. contributed samples and analytical tools; and R.K. and M.-L.H. supervised the project, designed research, interpreted, data and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for E.T. is Institute of Hematology, University of Perugia, Perugia, Italy. The current affiliation for V.B. is Institut für Rechtsmedizin, Ludwig Maximilians University, Munich, Germany.
Correspondence: Ralf Küppers, Institute of Cell Biology (Cancer Research), University of Duisburg-Essen Medical School, Virchowstrasse 173, 45122 Essen Germany; e-mail: ralf.kueppers@uk-essen.de.
References
Author notes
R.K. and M.-L.H. share last authorship.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal