Abstract
Abstract 932
Multiple myeloma (MM) is a heterogeneous disease. The discovery of a class of small non-coding RNAs (miRNAs) has revealed a new level of biological complexity underlying the regulation of gene expression. It may be possible to use this interesting new biology to improve our ability to risk stratify patients in the clinic.
We performed global miRNA expression profiling analysis of 163 primary tumors included in the UK Myeloma IX clinical trial. miRNA expression profiling was carried out using Affymetrix GeneChip miRNA 2.0; expression values for 847 hsa-miRNAs were extracted using Affymetrix miRNA QC tool and RMA-normalized. There are also 153 matching samples with gene expression profiles (GEP) and 72 matching cases with genotyping data available for integrative analyses. GEP was generated on Affymetrix HG-U133 Plus 2.0 and the expression values were RMA normalized; genotyping was performed on Affymetrix GeneChip Mapping 500K Array and the copy number values were obtained using GTYPE and dChip and were inferred against normal germ-line counterpart for each sample.
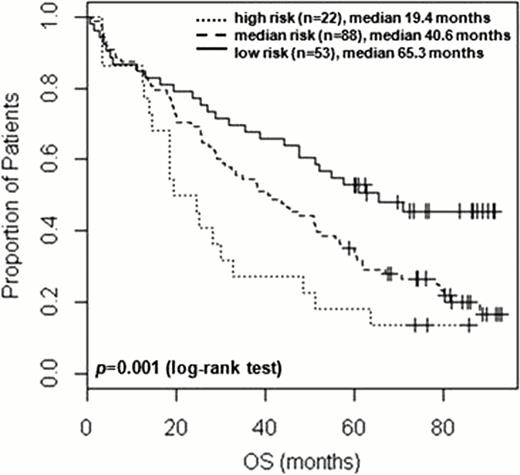
Firstly we have defined 8 miRNAs linked to 3 Translocation Cyclin D (TC) subtypes of MM with distinct prognoses, including miR-99b/let-7e/miR-125a upregulation and miR-150/miR-155/miR-34a upregulation in unfavourable 4p16 and MAF cases respectively as well as miR-1275 upregulation and miR-138 downregulation in favourable 11q13 cases. The expression levels of the miRNA cluster miR-99b/let-7e/miR-125a at 13q13 have been shown to be associated with shorter progression free survival in our dataset. Interestingly unsupervised hierarchical clustering analysis using these 8 miRNAs identified two subclusters among 11q13 cases, which have differential effect on overall survival (OS). We then evaluated the association of miRNA expression with OS and identified 3 significantly associated miRNAs (miR-17, miR-18 and miR-886-5p) after multiple testing corrections, either per se or in concerted fashion. We went on to develop an “outcome classifier” based on the expression of two miRNAs (miR-17 and miR-886-5p), which is able to stratify patients into three risk groups (median OS 19.4 months vs 40.6 months vs 65.3 months, log-rank test p = 0.001). The robustness of the miRNA-based classifier has been validated using 1000 bootstrap replications with an estimated error rate of 1.6%. The miRNA-stratified risk groups are independent from main adverse fluorescence in situ hybridization (FISH) abnormalities (1q gain, 17p deletion and t(4;14)), International Staging System (ISS) and Myeloma IX treatment arm (intensive or non-intensive).
Using the miRNA-based classifier in the context of ISS/FISH risk stratification showed that it can significantly improves the predictive power (likelihood-ratio test p = 0.0005) and this classifier is also independent from GEP-derived prognostic signatures including UAM, IFM and Myeloma IX 6-gene signature (p < 0.002). Integrative analyses didn't show enough evidence that the miRNAs comprising the classifier were deregulated via copy number changes; however, our data supported that the mir-17-92 cluster was activated by Myc and E2F3, highlighting the potential importance of Myc/E2F/miR-17-92 negative feedback loop in myeloma pathogenesis. We developed an approach to identify the putative targets of the OS-associated miRNAs and show that they regulate a large number of genes involved in MM biology such as proliferation, apoptosis, angiogenesis and drug resistance.
In this study we developed a simple miRNA-based classifier to stratify patients into three risk groups, which is independent from current prognostic approaches in MM such as ISS, FISH abnormalities and GEP-derived signatures. The miRNAs comprising the classifier are biologically relevant and have been shown to regulate a large number of genes involved in MM biology. This is the first report to show that miRNAs can be built into molecular diagnostic strategies for risk stratification in MM.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal