Abstract
Abstract 774
The presence of NPM1 mutations in the absence of FLT3 mutations typically confers favorable clinical responses and long term survival in acute myeloid leukemia patients. However, a subset of NPM1MUT:FLT3WT patients have extremely poor outcomes, indicating that uncharacterized genetic alterations influence the response of these patients. Identifying the causal alterations responsible for these poor prognoses would warrant more aggressive treatment strategies for these patients. While the impact of NPM1 mutations on AML is poorly understood, in vitro studies suggest NPM1 modulates p14ARF, resulting in p53 pathway dysregulation. The importance of p53 tumor suppressive functions are highlighted in AML patients with chromosome 17p deletions (containing the p53 locus) as they respond poorly to standard therapies. However, most AML patients possess wild type p53 alleles, suggesting that alterations to regulators of p53 activities may significantly influence ablation of the p53 pathway.
We currently have a limited knowledge of how p53 regulators effect AML progression and tumorigenesis in general. However, a recently described p53 and Mdm2 interacting protein, hnRNP K, has been shown to be a target of Mdm2-dependent degradation. This Mdm2 targeted degradation is thought to decrease p53-hnRNP K binding, resulting in aberrant p53-dependent transcriptional activities. Importantly, hnRNP K is aberrantly expressed in several human cancers, making it an attractive molecule for detailed examination of alterations in p53 pathway regulators. Thus, we are currently interrogating the relationship between hnRNP K and leukemic progression and are evaluating its potential as a prognostic marker and therapeutic target.
Using Mdm2-transgenic (Mdm2Tg) mice, we have demonstrated that hnRNP K levels decrease in the presence of excess Mdm2, indicating an in vivo relationship between hnRNP K and the Mdm2-p53 pathway. In order to directly examine the role of hnRNP K in cancer formation and leukemia, we generated hnRNP K knockout mice. Homozygous deletion of hnRNP K results in embryo lethality prior to embryonic day 13.5. Similarly, haploinsufficiency also frequently results in a neonatal lethal phenotype. However, nearly half of the hnRNP K+/− mice survive albeit exhibiting numerous developmental defects. Surviving hnRNP K+/− mice are tumor prone and some develop hematological neoplasms.
To examine the mechanisms by which hnRNP K influences the p53 pathway, we generated haploinsufficient hnRNP K mouse embryo fibroblasts (MEFs) and demonstrated that hnRNP K loss results in reduced p53 levels and diminished p53-dependent activation of p21. These results, coupled with our data from Mdm2Tg mice, establish an in vivo link between hnRNP K, the Mdm2-p53 pathway, and activation of p53 transcriptional targets.
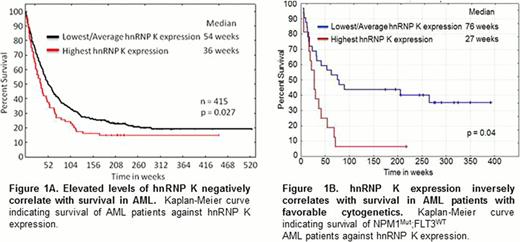
To explore the impact of hnRNP K expression in AML, we examined hnRNP K protein levels in 415 newly diagnosed AML patients. We observed that elevated hnRNP K levels associate with a significant increase in leukemic blasts and decreased survival (Figure 1A, p=0.027). Importantly, when assessed in the context of cytogenetically normal NPM1MUT:FLT3WT AML, high hnRNP K levels associate with poor outcomes within this a priori favorable prognostic group, with approximately 90% of these patients dying within 12 months (Figure 1B, p=0.049). These findings provide the first evidence that elevated hnRNP K levels associate with poor prognoses and suggest hnRNP K may represent a novel biomarker for risk assessment and a potential therapeutic target.
Currently, we have generated conditional hnRNP K knockout and transgenic alleles and are crossing them with Npm1+/− and Flt3IDT mice to directly examine its impact on leukemogenesis and the p53 pathway. Additionally, we are crystallizing hnRNP K in order to examine its interaction with Mdm2 and p53 for the purpose of evaluating its potential as a therapeutic target.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal