Abstract
Abstract 582
A clinical trial is being conducted at Stanford University for patients with mantle cell lymphoma (MCL) in first remission that interdigitates an autologous CpG-stimulated tumor cell vaccine with high dose therapy and autologous peripheral blood stem cell transplant (PBST). In this trial, patients receive a combination of PSCT and infustion of vaccine primed autologous T cells. Blood samples are collected before and after the priming vaccination and at regular time points following transplant, and they are assayed for minimal residual disease (MRD) and T cell repertoire by high-throughput sequencing of rearranged immunoglobulin heavy chain (IgH) and T cell receptor (TCR) genes.
We developed a sequencing-based platform, LymphoSIGHT, for MRD quantification in lymphoid malignancies. Using universal primer sets, we amplify rearranged IgH variable (V), diversity, and joining (J) gene segments from genomic DNA. To minimize the chance of somatic hypermutation interfering with detection of a cancer sequence, each IgH sequence is amplified by different sets of PCR primers in the three framework regions of the V segments and a common J segment primer. Amplified products can be sequenced to obtain >1 million reads and are analyzed using algorithms for clonotype determination. Tumor-specific clonotypes are identified for each patient based on their high frequency in the original tumor specimen. Quantitative MRD levels are then determined in serial samples of peripheral blood using spiked-in reference sequences. Our test has a sensitivity of 1 tumor cell per million leukocytes. This technology has also been extended to immunoglobulin light chain and T cell receptors. To quantify T cell responses to tumor vaccinations, the same samples are used for amplification, sequencing and analysis of the entire TCRB repertoire allowing for the assessment of T cell immune responses to the vaccine.
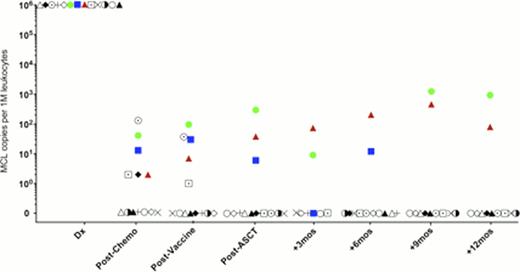
To date we have analyzed serial samples from 13 MCL patients who received the protocol of induction chemotherapy, vaccination, and PBST followed by vaccine primed T cell infusions and booster vaccinations. We have a pre-planned landmark analysis of MRD at 12 months post PBST. MRD was detected in blood samples immediately post-transplant in 3 patients, 2 of whom ultimately relapsed (Fig 1). In these two cases, disease was detected by sequencing 14 months and 4.5 months prior to detection by radiologic techniques. Clinical relapse in the second patient was originally restricted to the CNS; however, at a later point lymph node relapse occurred. The third patient has not shown signs of clinical relapse. The remaining 10 patients were MRD negative at 1 year following transplant, and these patients have not shown signs of clinical relapse with a median follow up of >24 months.
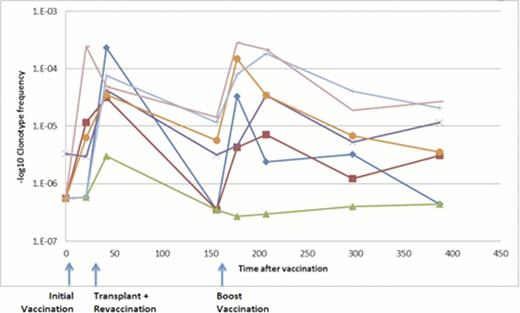
To identify T cells specific to the vaccination, we searched for clonotypes that were highly enriched (>10X) in an in vitro tumor-stimulated culture. In 2 of 3 patients assayed, we identified such clonotypes. The enrichment of these same clonotypes was also seen in the patients after vaccinations and boosters, adding to the evidence that they are directed against the vaccine. An example for one such patient is shown in Fig. 2. These clonotypes increased in frequency significantly after the boost by comparison to a set of frequency-matched unrelated clonotypes (p<2.5x 10−6). Figure 2 also shows the dynamics of these clonotypes through the course of treatment demonstrating frequency increases upon initial and booster vaccinations.
Our high-throughput sequencing method for MRD quantification shows promise for predicting clinical relapse in MCL patients after hematopoietic cell transplantation. This method provides detection lead-time that may be superior to clinical methods such as PET and MRI scanning but further follow up and greater numbers of patients will be required. Using the sequencing method, 77% of patients (10/13) were MRD negative at the landmark of 1 year post-transplant. Continued follow-up for molecular and clinical relapse is ongoing. T cell repertoire analysis identified clonotypes responding to the vaccination in some patients and follow up analyses will determine whether the presence of these clonotypes correlates with clinical outcomes in MCL patients.
Frequency and dynamics of vaccine-specific clonotypes over the course of treatment.
Frequency and dynamics of vaccine-specific clonotypes over the course of treatment.
Faham:Sequenta, Inc.: Employment, Equity Ownership, Research Funding. Carlton:Sequenta, Inc.: Employment, Equity Ownership, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal