Abstract
Abstract 3906
The clinical course of CLL patients ranges from an indolent and chronic disease to a rapidly progressing leukemia or lymphoma necessitating aggressive treatment. Very recent data have shown that genomic complexity evaluated by conventional karyotyping with DSP30/IL2 stimulation (Rigolin et al, Blood 2012) can be helpful in the prediction of the clinical course of patients with a normal fluorescence in situ hybridization (FISH) panel according to Dšhner et al (NEJM 2000). However, large studies investigating the impact of genomic complexity on the clinical course and possible correlations with other clinical parameters at time of diagnosis are still lacking. Therefore, we analyzed the role of genomic complexity in a large series of 329 CLL patients with available clinical data. Also, since SNP-array might soon represent an alternative to standard FISH in the clinical routine, we performed the so far largest comparison of FISH versus SNP-array in CLL.
Copy-number data, obtained using the Affymetrix Human Mapping GeneChip 6.0 arrays (SNP6), were derived from our previously reported CLL dataset (Rinaldi, Mian, Kwee et al, BJH 2011). The FISH panel interrogated deletions at 13q14.3, 11q22, 17p13 and trisomy 12. CLL diagnosis and management were based on the NCI Working group criteria (Hallek et al., Blood 2008). Real-time PCR on genomic DNA was performed to validate discordant FISH/SNP6 results.
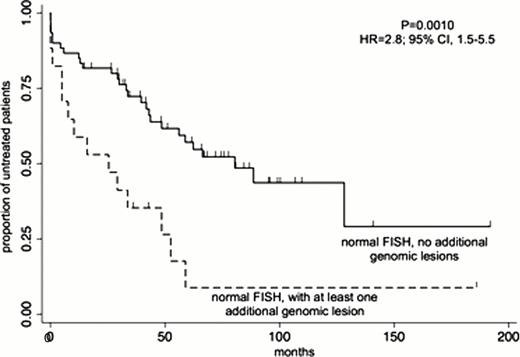
Seventy-seven of the 329 CLL patients (23%) presented a normal FISH. Among those, 17 patients (22%) had at least one large (>5 Mb) genomic aberration, different from those described by Dšhner et al (NEJM 2000). There was no correlation with the presence of TP53 mutations, since 13/13 were wtTP53. The DNA gains or losses did not occur at specific genomic loci, and their presence significantly affected the TTT (P= 0.0010; R=2.8; 95% CI, 1.5–5.5) (Figure 1), but not overall survival (OS) (p= 0.098; HR 2.3; 95% CI, 0.83–6.6). In a multivariate analysis including age, Binet stage, IGHV genes mutational status and the large genomic lesions, the latter three factors emerged as independent prognosticators for TTT with a significance of P<0.001, P<0.001 and P=0.036, respectively. In particular, the presence of at least one large genomic aberration identified patients with a shorter TTT among those with mutated IGHV genes (P= 0.0002; HR 5.0; 95% CI, 2.0–12.4) and with early stage disease (P= 0.005; HR 3.22; 95% CI, 1.36–7.62).
The concordance between FISH and SNP-array results was 93% for all analyzed genomic regions: 97% for trisomy 12, 96% for del 11q, 95% for del 17p and 84% for del 13q. False positive FISH results were observed for del 13q, while among the 18 cases with 17p loss by FISH half of the 10 cases with less than 40% of nuclei carrying the lesion were classified as normal by SNP6. We applied the FISH-based prognostic model developed by Dšhner et al (NEJM 2000) to our cohort classifying the 329 patients using results obtained by FISH or by SNP6: the Kaplan-Meier curves were comparable between FISH and SNP-array, both for OS as for TTT, and the log-rank test was highly significant for both approaches.
Patients with a normal FISH but with large genomic lesions detected by SNP-array have a worse TTT. As a whole, SNP-array has a high sensitivity and specificity and is able to identify the most important known prognostic genomic aberrations of CLL. A validation in prospective trials is needed.
Impact of at least one large genomic aberration on TTT in CLL patients with a normal FISH.
Impact of at least one large genomic aberration on TTT in CLL patients with a normal FISH.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal