Abstract
Abstract 3476
Most cases of Diamond Blackfan anemia are caused by haploinsufficiency for genes encoding proteins of the large or small ribosomal subunit. All of the ribosomal proteins affected in DBA are essential components of the ribosome required for the assembly of their respective subunits, including processing of the primary pre-rRNA transcript to mature 18S, 5.8S, and 28S rRNAs. Pre-rRNA processing signatures associated with ribosomal protein haploinsufficiency demonstrate a role for individual proteins in subunit assembly and can differ depending on which protein is affected. A facile pre-rRNA processing assay that can discriminate between loss of function alleles for different ribosomal protein genes would be an invaluable aide to DBA diagnosis and gene discovery efforts. Such an assay could also provide insight into different aspects of DBA pathophysiology.
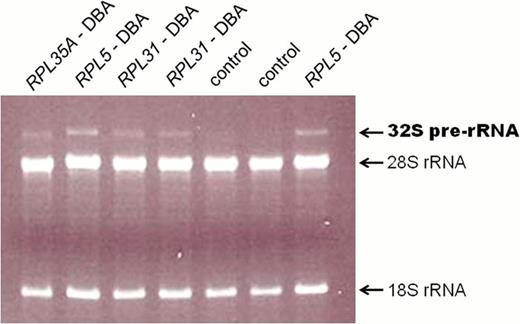
We have developed a robust procedure to assess pre-rRNA processing patterns in activated lymphocytes from the peripheral blood of patients with known or suspected Diamond Blackfan anemia. This assay typically involves the electrophoretic separation of total RNA from activated lymphocytes followed by Northern blotting with various hybridization probes to different rRNA precursors. Using this assay, we have found a common 32S pre-rRNA processing intermediate present in RNA from DBA patients with mutations in virtually all known large ribosomal subunit genes. This 32S pre-rRNA can be visualized in situ in gels stained with ethidium bromide (see accompanying figure) greatly simplifying the identification of large subunit ribosomal protein genes harboring loss of function mutations. As more and more ribosomal protein genes are identified within the DBA population, it has become increasingly important to distinguish between variants that affect ribosomal protein function and benign polymorphisms. Therefore, an analysis of pre-rRNA processing can be used to identify causative genes in patients with complex genotypes where sequence variants are found in more than one ribosomal protein gene. We analyzed a patient with variants in genes encoding RPS19 and RPL11, two known DBA genes, plus a deletion containing the RPL31 gene that has not been previously linked to DBA. Data analyses overwhelmingly support the deletion of RPL31 as the causative lesion in this patient and identify RPL31 as a new DBA gene. Analysis of pre-rRNA processing can also guide additional gene discovery efforts. We performed pre-rRNA processing studies on two patients lacking mutations in known DBA genes. In one case, we observed a clear defect in the18S rRNA pathway, implicating a gene involved in the biogenesis of the small ribosomal subunit, whereas in a second patient there is no evidence of a ribosome biogenesis defect suggesting that the underlying mutation may not affect ribosome synthesis. These results will help guide further efforts to identify causative genes in this patient cohort. Finally, we have used pre-rRNA processing patterns to begin to examine the mechanisms underlying remission in DBA patients. To date, we have examined two samples from DBA patients in remission and showed the ribosome synthesis defect is retained even while in remission for one of these patients, whereas the pre-rRNA processing defect has resolved in the other patient.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal