Abstract
Abstract 2960
Multiple myeloma (MM) is currently treatable but incurable. Many patients have a good initial response, but become refractory to therapy, and relapse. We propose that these patients, prior to therapy, harbor sub-populations of chemoresistant clones, which rely on intrinsic or environment-mediated drug resistance (EMDR). These sub-populations, initially a small fraction of the total tumor burden, are allowed to proliferate after aggressive therapy eradicates the sensitive clones.
We have previously proposed an approach, Adaptive Therapy (AT), which employs evolutionary principles to use the minimum amount and number of chemotherapeutic agents needed to reduce the tumor burden to a level that permits satisfactory quality of life, instead of high dose therapy aimed at eradication of the disease.
In this work we studied a cohort of 21 MM patients enrolled in an ongoing clinical trial using a single agent lenalidomide, combined with prednisone or dexamethasone in case of stable or progressive disease. We use a computational evolutionary model to determine the size, growth rate and drug resistance of sub-clones in each patient, and how these sub-populations evolve during therapy. We used these models to estimate progression of these patients for 48 months, as well as their hypothetical response to AT (Fig. 1).
Simulations using the model demonstrate that 9 patients would have benefited from AT, with lower accumulated dose and longer time to progression: 42.7mo vs. 15.4mo, P<0.001. These results indicate the need to develop a method to determine which patients benefit from this technique.
We propose an ex vivo approach to identify, prior to therapy, patients candidate for AT, and personalized agent combinations. This assay estimates the number, size, and drug resistance of sub-clones from live cells from MM patients' biopsies. We recreate within a 60uL microfluidic chamber, a stable drug gradient equivalent to a serial dilution in a 96-well plate. Each assay requires as few as 10,000 cells, which can be assayed in single or co-culture with stromal cells, to evaluate intrinsic and EMDR drug resistance levels. This assay was capable of determining the size and drug resistance properties of mixed cell line sub-populations at sizes as small as 10% (Fig. 2). We will next test this proof of principle with primary cells from relapsed patients, in order to determine the size and drug resistance properties of the MM clones present.
This preliminary work suggests that the standard aggressive therapy paradigm may not benefit all MM patients, but indeed accelerate relapse in patients with a pre-existing small sub-population of drug resistant clones. We also describe an approach that may be able to identify these patients.
Standard dose (MTD) versus AT for an IgD MM patient with initial serum level of 1,400mg/dl. Patient was treated with lenalidomide until month 6, when dexamethasone was added. (A) Serum m-protein level (black squares) was used to fit a computational model of the tumor burden. 50 possible solutions are shown as curves, and a validation data point is shown as a green square. (B) Each of the models was simulated under AT, the actual m-protein levels are shown for reference. (C) The simulation that best fit the validation data points shows that the majority of the burden was composed of lenalidomide-sensitive cells, which are eradicated after 4 months of treatment (red), allowing growth of a multidrug-resistant clone (orange). (D) AT prevents the growth of the MDR clones by maintaining the sensitive clone at a manageable level. (E) The models predict that under AT this patient would have a 95% of chance of being under the initial burden, compared to 5% under MTD, after 48 months.
Standard dose (MTD) versus AT for an IgD MM patient with initial serum level of 1,400mg/dl. Patient was treated with lenalidomide until month 6, when dexamethasone was added. (A) Serum m-protein level (black squares) was used to fit a computational model of the tumor burden. 50 possible solutions are shown as curves, and a validation data point is shown as a green square. (B) Each of the models was simulated under AT, the actual m-protein levels are shown for reference. (C) The simulation that best fit the validation data points shows that the majority of the burden was composed of lenalidomide-sensitive cells, which are eradicated after 4 months of treatment (red), allowing growth of a multidrug-resistant clone (orange). (D) AT prevents the growth of the MDR clones by maintaining the sensitive clone at a manageable level. (E) The models predict that under AT this patient would have a 95% of chance of being under the initial burden, compared to 5% under MTD, after 48 months.
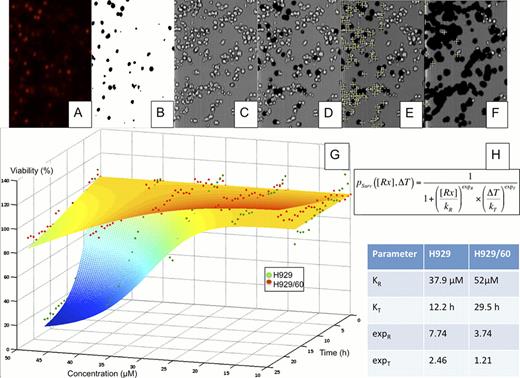
Drug resistance heterogeneity in MM. The human MM cell lines H929 and H929/60, resistant to HYD-1 were suspended in matrigel and loaded in a microfluidic chamber, across which a stable drug (HYD-1) gradient (from 0 to 50uM) was established. The media contained a fluorescent dye to mark the nuclei of dead cells (A). The fluorescent channel was converted to a black and white mask (B), which was superimposed to the brightfield channel (C), removing the dead cells (D). The number of live cells for each region of interest (ROI), or drug concentration, was quantified (E). After 24h of drug exposure the number of live cells is significantly reduced (F). Through non-linear regression (G), we determined the parameters of dose response of each population (H).
Drug resistance heterogeneity in MM. The human MM cell lines H929 and H929/60, resistant to HYD-1 were suspended in matrigel and loaded in a microfluidic chamber, across which a stable drug (HYD-1) gradient (from 0 to 50uM) was established. The media contained a fluorescent dye to mark the nuclei of dead cells (A). The fluorescent channel was converted to a black and white mask (B), which was superimposed to the brightfield channel (C), removing the dead cells (D). The number of live cells for each region of interest (ROI), or drug concentration, was quantified (E). After 24h of drug exposure the number of live cells is significantly reduced (F). Through non-linear regression (G), we determined the parameters of dose response of each population (H).
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal