Abstract
Abstract 2914
Based on clinical and biological data, a multistep model of disease progression starting in monoclonal gammopathy of undetermined significance (MGUS) continuing through MM, sometimes with an intermediate entity called smoldering MM (SMM), and ending in extramedullary disease, has been proposed.
To gain further insights into the role of the transcriptome deregulation in the transition from normal plasma cells (NPC) to clonal PC and from indolent clonal PC to malignant PC, we performed gene expression profiling (GEP) by Human Gene 1.0 ST Array (Affymetrix) in purified PC from 41 patients with MM, 33 with high-risk SMM and 20 with MGUS. In addition, 5 healthy donors were also included in order to relate the deregulation of GEP of clonal populations to normal condition.
Initially, we investigated whether the selected PC population from monoclonal gammopathy patients displayed specific GEP that were clearly distinguishable from NPC. 126 common genes were differentially expressed in MGUS, SMM and MM as compared to NPC (q-value < 10−5). The two most significant molecular and cellular functional categories were cell cycle and cell death (P < 0.01). The top 3 canonical pathways were EIF2 signaling, regulation of EIF4 and p70S6K signaling, and mTOR signaling (P < 0.001). Interestingly, 17 and 9 out of the 126 significant deregulated genes were small nucleolar RNA molecules and zinc finger proteins. GADD45A was the most significant up-regulated gene in clonal PC vs NPC.
Subsequently, we looked for genes deregulated in MGUS vs MM, SMM vs MM and MGUS vs SMM, in order to identify gene categories and molecular pathways involved in MM development. A total of 1,184 genes were differentially expressed between MGUS and MM (q-value < 10−5) with an imbalance in favor of antiapoptotic state in MM. Thus, BAG3, GADD45B, AKT1 and AKT2 were overexpressed in MM while APAF1 and BCL2L1 were underexpressed in MM. The molecular and cellular function most significantly affected in this comparison was cell death with 106 genes involved (P < 0.01). One of the top 10 genes upregulated in MM vs MGUS was TERC. In the SMM vs MM comparison, 1,163 genes were deregulated (q-value < 10−5). DNA replication, recombination and repair (ATM, RAD17, RAD50) was the most significant deregulated molecular and cellular function. Telomere extension by telomerase and Myc mediated apoptosis were included within the most significant canonical pathways. The analysis showed that a set of 627 differentially expressed genes was able to differentiate MGUS from SMM (q-value < 10−5). Cell death was identified as the most significant molecular function affected (P < 0.01). The RAC signaling pathway deregulation (P < 0.01) was included within the five most significant canonical pathways. In addition, NFKB signaling was also in the top canonical pathways (P < 0.01).
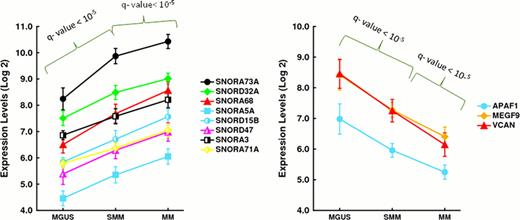
Finally, we also looked for those genes significantly deregulated among the three entities, which in turn were progressively up or downregulated from MGUS to SMM and to MM. We reasoned that these genes could be the most significant for promoting multistep transformation. Surprisingly, only eight out of the 2,974 significant genes exhibited a progressive deregulation (P < 0.0001) in the evolving stages of monoclonal gammopathies (Figure 1). All the genes with a progressive increase from MGUS to SMM and to MM were snoRNA. APAF1, VCAN and MEGF9 showed a progressive downregulation in the transition from MGUS to SMM and to MM.
Our data show that although MGUS, SMM and MM are not clearly distinguishable groups according to their GEP, several signaling pathways and genes were significant deregulated in the different steps of transformation process.
Expression of the genes significantly deregulated among the three entities.
Lahuerta:Janssen: Honoraria; Celgene: Honoraria. Oriol:Janssen: Honoraria; Celgene: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal