Abstract
Abstract 2593
NK-AML represents genetically heterogeneous group of disease. However genetic lesions affecting treatment outcome in patients with NK-AML are relatively unknown.
The discovery cohort consists of 67 NK-AML patients in complete remission (median age: 49.2, ranges: 19–70) without FLT-3 mutations. Genomic DNA was extracted from enriched AML cells at diagnosis or control specimens obtained after complete remission. Whole exomes were captured using Agilent SureSelect and sequencing were performed by HiSeq2000 with 41∼89× coverage. Bioinformatics analysis and identification of somatic mutation has been done by series of software such as BWA, Picard, GATK, VarScan 2, and custom-made scripts. All the data has been re-checked by manual inspection. Validation has been done independent set of cohort (358 NK-AML patients, median age: 51, ranges: 15–85) with Sanger sequencing on highly mutated target sites.
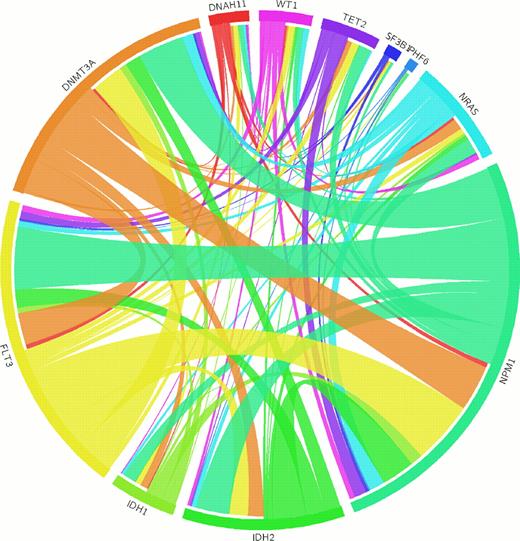
Filtering against dbSNP and COSMIC database generated 485 genes with somatic and structural variations. Among them, 41 genes were detected in more than two patients. In addition to well-known 28 mutations, 13 novel mutations with different frequencies were identified including genes responsible for structural maintenance of chromosome (SMC1A, 6.0%) and tumor suppressor function (FAT1, 6.0%). Most common type of mutation was missense mutation (70.8%), and substantial fraction of mutation was splicing site mutations (3.8%). The hematological system development and hematologic function were most highly enriched by the Ingenuity Pathway Analysis (IPA) as expected. CIRCOS plot analysis showed similar co-occurring pattern of recurrent mutations with previous reports. Hierarchical clustering analysis divided into four different groups according to the number of harboring mutations. In network analysis four distinct subgroups were observed ranging 21 to 3 gene network.
Using whole exome sequencing approach, a catalog of recurrent mutations was successfully defined in the patients with NK-AML without FLT3/ITD mutation. This candidate list of novel mutations should be tested further for therapeutic target and prognostic marker in the patients with NK-AML.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal