Abstract
Abstract 2549
BCR-ABL1 mutation testing is recommended for CML and Ph+ ALL patients who fail first line tyrosine kinase inhibitor (TKI) therapy or who have a suboptimal response to therapy. BCR-ABL1 mutations in the kinase domain (KD) of ABL1 account for at least 40–50% of all TKI resistant cases. Rare mutations such as E123Q and T212R in the regulatory domain of ABL upstream of the kinase domain have also been reported to lead to resistance to imatinib. The current gold standard for BCR-ABL1 mutation detection is Sanger sequencing, which has an analytical sensitivity of ∼10–30%. Based on recent findings that mass spectrometry can identify low level BCR-ABL1 mutations that confer clinical resistance in patients sooner than Sanger sequencing, it is likely useful to have a significantly more sensitive BCR-ABL1 test than Sanger sequencing. Although commercial NGS cancer panels have included ABL1 in the region of interest, ABL1 resistance mutations should be sequenced from BCR-ABL1 fusion transcripts instead of being sequenced from genomic DNA as in the commercial panels. Here we developed a fusion transcript based BCR-ABL1 mutation assay on the scalable and cost-effective Ion Torrent platform that has 1–5% sensitivity and comprehensive coverage of the kinase domain, regulatory domain, and the SH2/SH3 domains. The assay was designed to detect both the major and minor BCR-ABL1 fusion gene products and can also detect the micro BCR-ABL1 fusion product accounting for over 99% of all CML and Ph+ ALL patients.
RT and long range PCR was performed to amplify BCR-ABL1 e1, e13, and e14 fusion transcripts and the PCR products were enzymatically fragmented and ligated with Ion Torrent sequencing adaptors. Size-selected libraries were quantified, pooled, amplified with OneTouch system and sequenced with Ion Torrent PGM. Sequencing data was analyzed with Torrent Suite 2.2 and the associated variant caller with variant frequency cutoff adjusted to 1%.
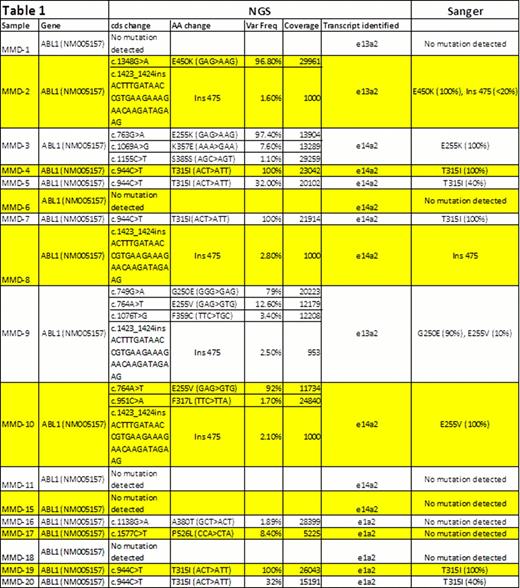
Initial work with cell lines harboring the T315I mutation in both e1 and e14 BCR-ABL1 transcript types diluted into wild type cell line demonstrated that Ion Torrent NGS can detect T315I at least down to 1%. In a set of 17 blinded clinical samples, Ion Torrent NGS not only identified all the mutations found by Sanger sequencing but additionally identified rare imatinib resistant mutations such as K357E (MMD-3) present at 7.6%; this mutation was previously reported in patient CD34+CD38-stem cells (Table 1). This patient also expressed the E255K resistance mutation; in patients, the presence of multiple resistance mutations has been shown to be an important predictor of poor response.
Understanding whether compound mutations are present in cis or in trans may be important in understanding therapy resistance. For patient MMD-9, although both the predominant mutation G250E (79%) and subclone E255V(12.6%) were identified by both Sanger sequencing and NGS, only Ion Torrent was able to show that the two mutations were on different reads, indicating that the mutations are on different alleles. The BCR-ABL1 Ion Torrent based assay reads DNA fragments between 150–200bps in size and can identify cis and trans mutations from individual fragments with this read length, which is not possible by Sanger sequencing. In MMD-9, another low frequency subclone F359C (3%) was detected by only NGS and may have important implications due to its reported sensitivity to dasatinib and not imatinib or nilotinib. Similarly, in MMD-10 the E255V predominant mutation was identified by both NGS and Sanger sequencing, while the F317L mutation with a low frequency of 1.70% was solely detected by NGS. In this patient, a combination of dasatinib and nilotinib treatment may be required to eliminate the dasatinib sensitive E255V dominant clone and F317L nilotinib sensitive subclone. Ion Torrent NGS was also capable of identifying and calling the 35 base pair 475 insertion from 4 samples (MMD-2 and 8–10), only two of which were detected by Sanger sequencing.
In our ongoing study, the low-level mutations not detected in Sanger sequencing will be confirmed with analyses on the MiSeq platform and mutation enrichment methods. Additional patient samples representing 30 CML patients will be analyzed and presented.
Wong:MolecularMD: Equity Ownership.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal