Abstract
Abstract 2526
Single nucleotide polymorphism (SNP) is an inter-individual genetic variation which could explain inter-individual differences of response/survival to chemotherapy. The present study was attempted to build up risk model of survival for acute myeloid leukemia (AML) patients with normal karyotype (AML-NK).
A total of 247 patients with AML-NK was included into the study. Genome-wide SNP array (Affymetrix SNP-array 6.0) was performed in the discovery set (n=118), and genotypes were analyzed for overall survival (OS). After identifying significant SNPs for OS in single SNP analyses, risk model was constructed. Replication was performed in an independent validation cohort (n=129).
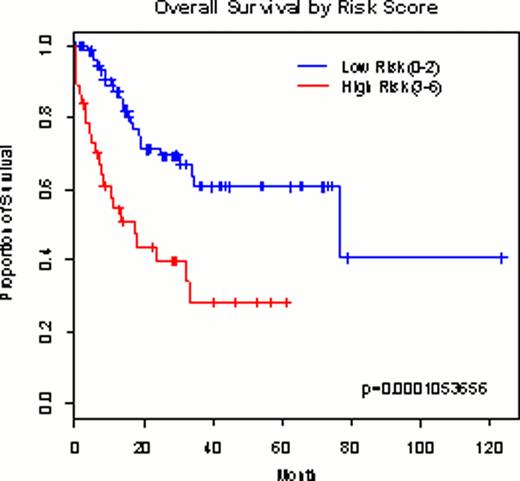
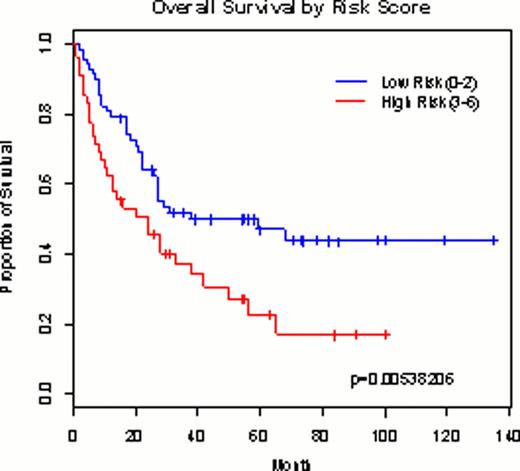
Out of 632,957 autosomal SNPs meeting genotype data filtration criteria, a total of 82 SNPs were selected and passed into the next step of validation in an independent cohort. In the risk model generation step, finally 4 SNPs (rs2826063, rs12791420, rs11623492 and rs2575369) were meeting stringent criteria for SNP selection as follows: 1) p-value < 0.10 from Cox proportional hazards regression model in adjustment with age and WBC counts at diagnosis; 2) minor allele frequency > 0.05; 3) call rate > 95.0%; 4) high linkage disequilibrium r2 < 0.8. These 4 SNPs were introduced into the risk model, and patients was grouped into 2 groups according to the number of deleterious variables including 4 SNPs and 2 clinical variables (i.e. age and WBC counts at presentation): risk score 0–2 as a low risk (n=80) and 3–6 as a high risk (n=38). The risk model could stratify the patients according to their OS in discovery (p=1.053656•10−4) and in validation set (p=5.38206•10−3). The risk model showed a higher AUC than those being incorporated only clinical or only 4 SNPs, suggesting improved prognostic stratification power of combined model.
Genome-wide SNP based risk model obtained from 247 patients with AML-NK can identify high risk group of patients with poor survival using genome wide SNP data. (Clinicaltrials.govIdentifier:NCT01066338)
Survival curve according to the predictive risk model in the discovery cohort (n=118; A) and validation cohort (n=129; B).
Survival curve according to the predictive risk model in the discovery cohort (n=118; A) and validation cohort (n=129; B).
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal