Abstract
Abstract 1717
Aberrant DNA methylation of tumor suppressor genes is a common event in myelodysplastic syndromes (MDS) and acute myeloid leukemias (AML). Therefore hypomethylating agents like azacytidine (AZA) and decitabine seem to be a good therapeutic approach for the treatment of these diseases. Clinical experience and recent published data have demonstrated that AZA is effective for the treatment of MDS and AML patients. However, the prognostic impact of the aberrant hypermethylation on response and outcome to AZA treatment remains to be determined. For this reason the influence of the methylation status of a selected set of tumor suppressor genes on the overall survival and clinical response in MDS and AML patients, prior to treatment with AZA was studied.
A total of 78 patients with MDS or AML who had been treated with AZA were evaluated. Among these, 25 were excluded because they had received less than 4 cycles, AZA was used after allogeneic stem cell transplantation, response was not assessable, or there was not enough quality DNA available. So finally, the study was focused in 53 patients: 36 MDS and 17 AML. Most of the AML included in the study had low blast count (20–30%). Responses were assessed according to the IWG MDS criteria in accordance to Fenaux et.al, and IWG AML criteria following European LeukemiaNet recommendations. DNA methylation status was analyzed using the Methylation Specific Multiplex Ligation Probe Amplification (MS-MLPA), with a panel of 24 different tumor suppressor genes related to cell cycle control, apoptosis regulation, DNA repair, cell adhesion and cell growth.
In the study cohort 47% of patients had cytogenetic alterations prior to AZA treatment, 4 with isolated -5/del(5q), 7 with isolated −7/del(7q), 4 with trisomy 8, 4 with not isolated -5/del(5q), 1 with trisomy 14, and 5 with complex cytogenetics. Methylation analysis showed that most patients (74%) had at least one methylated gene, but only 10% of patients displayed more than 3 methylated genes. The most frequently methylated genes were IGSF4 (28.3%), CDKN2B (24.5%), ESR1 (22.6%), CDH13 (17%) and CDKN1B (11.3%).
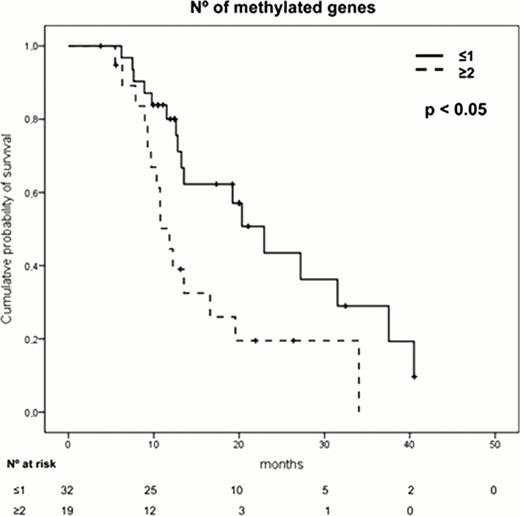
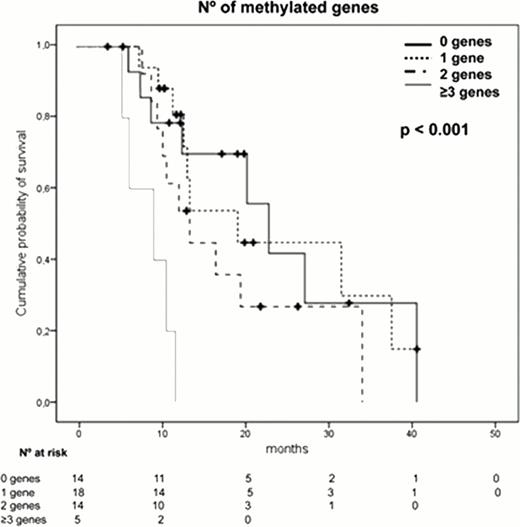
The presence of a high number (≥2) of methylated genes (p=0.02), an adverse cytogenetics (p=0.03) or anemia (p=0.04) were independent prognostic factors associated with shorter overall survival. Moreover, the analysis of those patients displaying “no methylation”, patients with 1 methylated gene, patients with 2 methylated genes and those with more than 3 methylated genes, showed that as the number of methylated genes increases, the survival was shorter. The patients displaying the highest level of methylation (more than 3 genes), had a very short survival (median OS of 9.3 months, p<0.001).
Forty-nine per cent of all patients responded to azacytidine. The statistical analysis revealed that the number of methylated genes did not correlate in general with the clinical response to AZA, although four of the five patients with more than 3 genes methylated did not respond. By contrast, cytogenetics was the only factor that independently influenced the response to AZA. Thus 64.7% of patients with favorable cytogenetics responded to AZA, while only 28.6% of those with adverse cytogenetics responded (p=0.03). Interestingly, our study included 3 MDS patients with isolated chromosome 7 alterations, and all of them responded.
In summary, these results suggest that the analysis of the methylation status before AZA treatment by a feasible methodology such as MS-MLPA could be used in a clinical setting in order to identify a group of patients with poor survival, in which other alternative therapeutic approaches might be considered.
Ramos:Celgene, Novartis, Amgen, Pfizer, and Merck Sharp & Dohme: Honoraria. Hernández:Celgene: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal