Abstract
High expression of several genes have been found to be prognostically relevant in AML, such as EVI1, BAALC, MN1 and ERG [1–4]. These studies have mostly reported cutpoints at expression level quartiles with the highest prognostic value. Translation to the clinic remains difficult because a quartile definition cannot be applied to an individual patient. Therefore, from a clinical practice point, standardization has remained an important objective.
Using a standardized microarray for gene expression profiling we set out to develop and validate expression cutpoints for the EVI1, BAALC, MN1 and ERG genes that best predict overall survival (OS) for intermediate cytogenetic risk cases, rather than normal karyotype.
For training purposes, we employed a cohort of 147 intermediate cytogenetic risk cases [5, 6], and an external dataset of 242 cases [7]. For validation purposes, a cohort of 215 intermediate cytogenetic risk cases was employed [6]. Expression of the EVI1, BAALC, MN1 and ERG genes was measured using a custom microarray (AMLprofiler) with IVD-grade reagents and Affymetrix DX2 equipment.
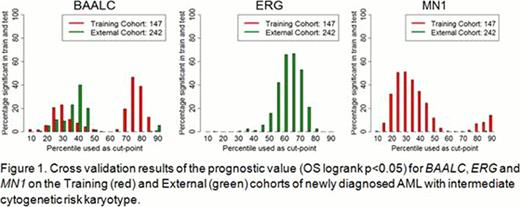
For the BAALC, MN1 and ERG genes, cutpoints were considered from the 10th to 90th percentile in steps of 5%. Optimal cutpoints were selected by performing 100-fold cross validations of the expression data of the training cohort and external cohort. EVI1 has a discontinuous distribution of expression [1]. Because of the low prevalence of high-EVI1 in AML we did not perform cross validation but used the entire training cohort to derive the best cutpoint.
For BAALC the 30th percentile cutpoint was verified to be clinically relevant in the training and external cohorts (Figure 1). ERG had no significant splits in the training data, and around the 65th percentile in the external cohort (Figure 1). MN1 had a broad range of significant splits around its 30th expression percentile in the training cohort but none in the external cohort (Figure 1). Due to the lack of consistent results across datasets, we have not attempted a validation of ERG and MN1. The best EVI1 cutpoint in the training set was found to be the 6th percentile. This cutpoint turned out to be concordant with a previously developed Q-PCR cutpoint [8, 9].
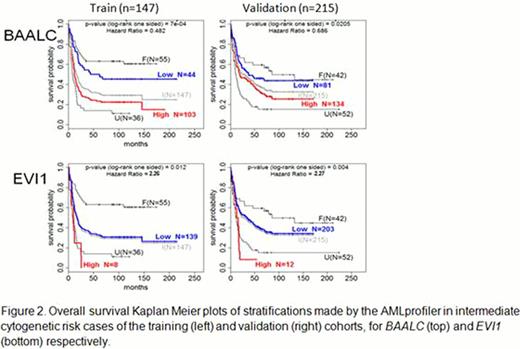
Validation of the BAALC and EVI1 cutpoints on the independent validation cohort of intermediate cytogenetic risk cases resulted in significantly more favorable OS for cases with low-BAALC expression (HR 0.686, p=0.0205, Figure 2) and worse OS for cases with high-EVI1 expression (HR 2.27, p=0.004, Figure 2).
In a multivariable analysis, including age, gender, white blood cell count and, blast percentage in BM, high-EVI1 retained independent prognostic value [HR 2.39, p=.008] while low-BAALC retained independent prognostic value only within the group of low-EVI1 cases [HR 0.602, p=.015].
For BAALC and EVI1, but not for MN1 and ERG, clinically relevant cutpoints have been developed that remain independent prognostic factors in intermediate cytogenetic risk AML and which through their incorporation in the AMLprofiler assay can be used for standardized measurements across different laboratories.
van Beers:skyline diagnostics: Employment, Equity Ownership, Patents & Royalties. Brand:skyline diagnostics: Employment, Equity Ownership. van Vliet:skyline diagnostics: Employment, Equity Ownership. Vietor:skyline diagnostics: CEO skyline diagnostics Other, Equity Ownership. Valk:skyline diagnostics: Equity Ownership. Lowenberg:skyline diagnostics: CSO skyline diagnostics Other, Equity Ownership.

This icon denotes a clinically relevant abstract
Author notes
Asterisk with author names denotes non-ASH members.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal