Abstract
Abstract 112
Alternative splicing of RNA is a fundamental biological process that affects almost all multi-exonic genes to promote protein diversity. Of the 3 modes of RNA splicing that also include exon skipping and alternative splice site usage, intron retention (IR) is the least abundant and least understood. Despite isolated instances of IR-associated biological function, IR has been widely regarded as a failure in the splicing machinery to excise intronic sequences from pre-messenger RNAs. Since an overall role for IR is unknown, we systematically examined the impact of IR in normal primary myeloid cells during differentiation. Using messenger RNA sequencing (mRNA-seq) and a novel algorithm we termed IRFinder, we determined intron-retaining genes that were differentially regulated in FACS-purified cells at three progressive stages of mouse granulopoiesis; CD34+Kit+Gr-1low promyelocytes, CD34−Kit−Gr-1mid myelocytes and CD34−Kit−Gr-1high granulocytes. We demonstrated that IR exhibits a specific pattern of dynamic regulation in 86 genes during granulocytic differentiation.
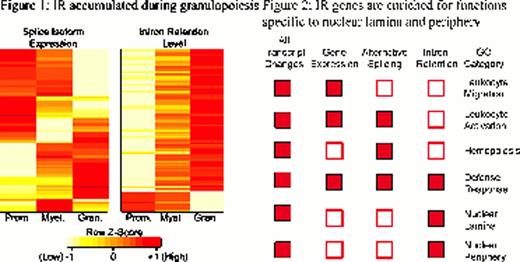
intron retaining transcripts were predominantly accumulated during differentiation, with low levels in promyelocytes, increasing through myelocytes to the highest levels in granulocytes. These genes include those with functions specific to granulocytes (Lyz2 and MMP8) and those governing the nuclear architecture (Lmnb1, Lmnb2, Lbr and Npm1), consistent with the unique change of nuclear morphology from promyelocytes to granulocytes. Figure 2, IR was significantly associated with nuclear localisation and functions involving the nuclear periphery. Subsequent mRNA-seq and IRFinder analysis of FACS purified human granulocytes displayed significant overlap of intron retaining genes between human and mouse (71/86 genes, P=2.85E-22, hypergeometric test), showing that IR is conserved between these two species. Inhibition of nonsense-mediated decay (NMD) in primary granulocytes using caffeine coupled with actinomycin D resulted in marked accumulation of 39/86 intron retaining transcripts (P<0.05, RUV procedure with Holm-Bonferroni correction), indicating that intron retaining transcripts are subjected to degradation by NMD. Mass spectometry analysis detected proteins encoded by 21 NMD-regulated intron-retaining genes. There was a strong negative correlation of protein expression with levels of IR (P=0.0015, binomial test). In mouse bone marrow reconstitution experiments, we showed that enforced re-expression of the Lmnb1 gene, which displayed the highest levels of endogenous IR led to decreased granulocyte cell count, increased nuclear volume by 30% and altered nuclear morphology. We conclude that IR coupled with NMD is a conserved physiological mechanism that may provide an energetically favourable level of gene expression control during granulopoiesis. Our findings establish a foundation to examine the role of IR- coupled NMD in normal haemopoiesis as well as in hemopoietic diseases now known to be affected by mutations of splicing factors.
intron retaining transcripts were predominantly accumulated during differentiation, with low levels in promyelocytes, increasing through myelocytes to the highest levels in granulocytes. These genes include those with functions specific to granulocytes (Lyz2 and MMP8) and those governing the nuclear architecture (Lmnb1, Lmnb2, Lbr and Npm1), consistent with the unique change of nuclear morphology from promyelocytes to granulocytes. Figure 2, IR was significantly associated with nuclear localisation and functions involving the nuclear periphery. Subsequent mRNA-seq and IRFinder analysis of FACS purified human granulocytes displayed significant overlap of intron retaining genes between human and mouse (71/86 genes, P=2.85E-22, hypergeometric test), showing that IR is conserved between these two species. Inhibition of nonsense-mediated decay (NMD) in primary granulocytes using caffeine coupled with actinomycin D resulted in marked accumulation of 39/86 intron retaining transcripts (P<0.05, RUV procedure with Holm-Bonferroni correction), indicating that intron retaining transcripts are subjected to degradation by NMD. Mass spectometry analysis detected proteins encoded by 21 NMD-regulated intron-retaining genes. There was a strong negative correlation of protein expression with levels of IR (P=0.0015, binomial test). In mouse bone marrow reconstitution experiments, we showed that enforced re-expression of the Lmnb1 gene, which displayed the highest levels of endogenous IR led to decreased granulocyte cell count, increased nuclear volume by 30% and altered nuclear morphology. We conclude that IR coupled with NMD is a conserved physiological mechanism that may provide an energetically favourable level of gene expression control during granulopoiesis. Our findings establish a foundation to examine the role of IR- coupled NMD in normal haemopoiesis as well as in hemopoietic diseases now known to be affected by mutations of splicing factors.
Rasko:Genea Ltd: Employment; Rarecyte: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal