Abstract
Sézary syndrome (SS) is an aggressive cutaneous T-cell lymphoma (CTCL) of unknown etiology in which malignant cells circulate in the peripheral blood. To identify viral elements, gene fusions, and gene expression patterns associated with this lymphoma, flow cytometry was used to obtain matched pure populations of malignant Sézary cells (SCs) versus nonmalignant CD4+ T cells from 3 patients for whole transcriptome, paired-end sequencing with an average depth of 112 million reads per sample. Pathway analysis of differentially expressed genes identified mis-regulation of PI3K/Akt, TGFβ, and NF-κB pathways as well as T-cell receptor signaling. Bioinformatic analysis did not detect either nonhuman transcripts to support a viral etiology of SS or recurrently expressed gene fusions, but it did identify 21 SC-associated annotated long noncoding RNAs (lncRNAs). Transcriptome assembly by multiple algorithms identified 13 differentially expressed unannotated transcripts termed Sézary cell-associated transcripts (SeCATs) that include 12 predicted lncRNAs and a novel transcript with coding potential. High-throughput sequencing targeting the 3′ end of polyadenylated transcripts in archived tumors from 24 additional patients with tumor-stage CTCL confirmed the differential expression of SC-associated lncRNAs and SeCATs in CTCL. Our findings characterize the SS transcriptome and support recent reports that implicate lncRNA dysregulation in human malignancies.
Introduction
Mycosis fungoides (MF) and Sézary syndrome (SS) are the most common forms of cutaneous T-cell lymphoma (CTCL). SS, also referred to as the leukemic phase of erythrodermic CTCL, is caused by a malignant clonal proliferation of central memory T cells that leads to erythroderma, lymphadenopathy, malignant cells in the peripheral circulation, and immune dysfunction. The current armamentarium of chemotherapeutic and biologic agents is able to palliate but not cure this aggressive non-Hodgkin lymphoma, which has a median survival of 2-4 years.1 The pathogenic mechanisms underlying CTCL are poorly understood, and improved genomic characterization of this disease may shed light on both disease mechanisms and previously unexplored therapeutic targets.
RNA-Seq can be used to obtain global cell transcriptome profiles and thus represents a powerful discovery tool in cancer biology.2 A major advantage of RNA-Seq over conventional DNA microarray analyses is the ability to identify previously undescribed transcripts, such as long noncoding RNAs (lncRNAs). This class of genes is transcribed but not translated and can modulate cellular processes such as epigenetic gene regulation, cell cycle control, and apoptosis.2–4 Furthermore, RNA-Seq can identify nonhuman transcripts, such as those of viral origin, to facilitate the search for potential infectious causes of human diseases.
Here, we have used RNA-Seq to define the transcriptome of pure, freshly sorted populations of Sézary cells (SCs) and patient-matched CD4+ T cells in 3 patients with SS. We did not find evidence to support a viral cause of SS or recurrent gene fusions. Analysis of the protein-coding gene expression signature shared by all 3 patients confirmed dysregulation of several key cancer pathways, including those involving phosphatidylinositol 3-kinase (PI3K), nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB), and transforming growth factor β (TGFβ). In addition to coding genes, 21 annotated SC-associated lncRNAs were differentially expressed in SS. Transcriptome assembly by multiple algorithms further identified 13 previously unannotated and undescribed Sézary cell–associated transcripts (SeCATs) that are differentially expressed in SCs from all 3 patients with SS. Twelve SeCATs are predicted to be noncoding, and 1 unexpectedly displays protein-coding potential conserved with nonhuman primates. These newly identified SeCATs showed modest evolutionary conservation and high tissue specificity, suggesting a potential functional role in T cells. High-throughput sequencing of formalin-fixed, paraffin-embedded (FFPE) tumors from 24 patients with stage IIB/III MF showed differential expression of SC-associated lncRNAs and SeCATs in tumor-stage MF as well as SS. These data identify candidate lncRNAs with potential roles in the pathogenesis of cancer.
Methods
Patients
Following informed consent per the Declaration of Helsinki, Sézary patient samples were collected under a protocol approved by the Institutional Review Board at Stanford University Medical Center. All patients had SS by revised staging criteria5 with clinical stage IVA disease. Patient characteristics are described in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The 24 MF tumor samples were collected between 1989 and 2008 and fall under exemption 4. All samples were obtained from patients with either clinical stage IIB or III CTCL. All diagnoses were confirmed by a board-certified dermatopathologist.
Cell sorting
PBMCs were prepared by Ficoll-Hypaque density-gradient centrifugation. PBMCs were stained with fluorochrome-labeled anti–human monoclonal antibodies (Biolegend Inc) to CD45 (clone HI30), CD4 (clone RPA-T4), and CD3 (clone HIT3a). T-cell receptor (TCR) Vβ clonality was determined with the TCR Vβ Repertoire Kit (Beckman-Coulter). Antibody-stained patient lymphocytes were sorted into CD3+/CD4+/Vβ+ and CD3+/CD4+/Vβ− fractions with the use of an Influx flow cytometer (Becton Dickinson).6
Cells and cell lines
Human CTCL lines MyLa and SeAx were generous gifts from Dr K. Kalthoft (Aarhus University, Denmark) and were cultured in RPMI with 10% FBS and 200 IU/mL IL-2.7,8 Hut-78 cells were obtained from American Type Culture Collection. Normal CD4+ T cells were obtained from healthy volunteers from the Stanford Blood Center; CD4+ T cells were prepared by Ficoll-Hypaque density-gradient centrifugation, followed by CD4+ selection with the use of CD4+ microbeads (Miltenyi Biotec).
RNA isolation
Total RNA was isolated from sorted CD3+/CD4+/Vβ+ and CD3+/CD4+/Vβ− lymphocytes (∼ 2-4 × 106) with the use of Trizol (Invitrogen), followed by DNA removal with the TURBO DNA-free kit (Ambion). RNA integrity was verified with an Agilent 2100 Bioanalyzer. Total RNA from cell lines was prepared with the RNeasy Plus kit (QIAGEN). Total RNA extraction from FFPE was performed as previously described.9 Briefly, multiple 20-μm sections were deparaffinized, subjected to protease digestion, and then nucleic acid extracted with the use of the RecoverAll Total Nucleic Acid Isolation Kit (Ambion).
Library preparation and sequencing
RNA-Seq libraries were prepared with the mRNA Seq Sample Prep Kit (Illumina). mRNA was isolated by polyA selection from 1 to 2 μg of total RNA, fragmented, and randomly primed for reverse transcription, followed by second-strand cDNA synthesis. After end repair, adenylation of 3′ ends, adapter ligation, isolation of ∼ 200-bp (patient 1) or ∼ 300-bp (patients 2 and 3) cDNA fragments and subsequent PCR amplification, 50-bp (patient 1) or 101-bp (patients 2 and 3) paired-end sequencing reads were obtained with the Illumina HiSeq platform. 3SEQ (3′ end of polyadenylated transcripts) was performed as described previously.9 Briefly, oligo-dT–directed reverse transcription generated cDNAs corresponding to 3′ ends of polyadenylated transcripts; after linker ligation, size selection, and PCR amplification, the cDNAs were subjected to deep sequencing on the Illumina GAIIx platform with a raw read length of 36 bp.
Sequence analysis
RNA-Seq reads were aligned to the human reference sequence National Center for Biotechnology Information (NCBI) build 36.1/hg1810 with TopHat.11 Ab initio assembly was performed with Cufflinks12 and analyzed with Cuffdiff to determine differential expression. RefSeq and Gencode databases were used as reference annotations to calculate values of fragments per kilobase of transcript per million mapped reads for known transcripts. Scripture13 and Trinity14 were used for de novo transcriptome assembly. Scripture assembly was used to categorize transcripts as protein-coding, noncoding, pseudogene, or novel on the basis of a > 1-bp intersection with annotated transcripts from University of California Santa Cruz (UCSC),15,16 Gencode,17 RefSeq,18 and Ensembl.19 3SEQ reads were aligned to hg18 with Bowtie.20 Read counts for the 3′-most exon of each gene were calculated with a self-developed script (K.Q.). Further details of sequence analysis are described in supplemental Methods.
Analysis of tissue specificity
Two publicly available RNA-seq datasets from 16 normal human tissues (ArrayExpress no. E-MTAB-513 and Gene Expression Omnibus no. GSE30554) were used to determine the relative expression of SC-associated lncRNAs and SeCATs.21 Further description of these datasets and the analysis performed can be found in supplemental Methods. Relative expression was normalized to average expression in CD3+/CD4+/Vβ− lymphocytes.
Conservation analysis
GSEA
Gene set enrichment analysis (GSEA) was performed with a permutation of 1000. Normalized enrichment scores (NESs) were calculated to account for differences in gene set size as well as correlations between our dataset and other gene signatures. The false discovery rate (FDR) q value represents the probability of a false-positive finding for a given NES. FDR q value thresholds of approximately 0.25 are recommended when phenotype permutations are performed.25
Viral sequence detection
The RINS algorithm was used with default parameters to analyze RNA-Seq data from each patient's SCs as well as control lymphocytes.26
Fusion transcript discovery
The DeFuse algorithm (Version 0.4.0) was used with default parameters to analyze RNA-Seq data from each patient's SCs as well as control lymphocytes.27 PCR from cDNA was used to test for the presence of predicted in-frame gene fusions. ChimeraScan was also used to analyze RNA-Seq data with default parameters.28
cDNA synthesis and PCR
cDNA was synthesized from DNase-treated total RNA using the iScript cDNA synthesis kit (Bio-Rad). PCR amplification was performed with the Phusion High-Fidelity PCR kit (New England Biolabs).
Quantitative RT-PCR analysis
Quantitative PCR (qPCR) was performed with the Maxima SYBR Green qPCR master mix (Fermentas) and the Stratagene Mx3000P (Agilent Technologies) thermocycler. Samples were run in triplicate and normalized to glyceraldehyde 3-phosphate dehydrogenase. Relative mRNA expression was calculated with the ΔCT method. Primer sequences are listed in supplemental Table 1.
Results
RNA-Seq defines a global set of transcriptional aberrations in SS
To characterize RNA expression in SS, we performed paired-end, high-throughput sequencing with the Illumina HiSeq platform on malignant SCs as well as patient-matched, normal CD4+ T lymphocytes purified by flow cytometry directly from the peripheral blood of 3 unrelated persons with SS (Figure 1A). SCs were defined as CD3+/CD4+ cells with clonal Vβ expression, whereas patient-matched, CD3+/CD4+ cells with polyclonal Vβ expression isolated in parallel were used as control. We obtained an average of 112 million reads for each sample, > 90% of which aligned to the human genome (supplemental Table 2). This depth of coverage is comparable with recent data in human B cells demonstrating detection of 90% of transcripts.29 Clonal mRNA expression of the specific malignancy-associated Vβ was seen in sorted malignant populations, compared with polyclonal expression of a diversity of Vβ mRNAs in control lymphocytes (Figure 1B), confirming the high degree of purity of malignant and nonmalignant cells in each of the populations analyzed.
Analysis of transcriptome data in SS. (A) Schematic of experimental design. SC and patient-matched normal CD4+ T lymphocytes were purified by flow cytometry, with SC sorted using Vβ antibodies to the malignant clone, followed by paired-end RNA-Seq. Sequence reads underwent genome alignment followed by assembly into gene transcripts and determination of differential expression. Scripture was used as an orthogonal means of transcriptome assembly, while both Scripture and Trinity were used for novel transcript discovery. (B) RNA-Seq tracks over the T-cell receptor beta locus, with normalized read scale between each SC and normal matched CD4+ T-cell control. Clonal Vβ transcript peaks are off-scale in SC, with read number shown next to each. (C) Transcript distribution by category in SS. Transcripts were annotated using a conglomeration of UCSC, Gencode, RefSeq, and Ensembl as reference. An FPKM > 1.5 was required in either SC or control cells. (D) Differentially expressed protein-coding, noncoding, pseudogene, and unannotated novel transcripts in SS.
Analysis of transcriptome data in SS. (A) Schematic of experimental design. SC and patient-matched normal CD4+ T lymphocytes were purified by flow cytometry, with SC sorted using Vβ antibodies to the malignant clone, followed by paired-end RNA-Seq. Sequence reads underwent genome alignment followed by assembly into gene transcripts and determination of differential expression. Scripture was used as an orthogonal means of transcriptome assembly, while both Scripture and Trinity were used for novel transcript discovery. (B) RNA-Seq tracks over the T-cell receptor beta locus, with normalized read scale between each SC and normal matched CD4+ T-cell control. Clonal Vβ transcript peaks are off-scale in SC, with read number shown next to each. (C) Transcript distribution by category in SS. Transcripts were annotated using a conglomeration of UCSC, Gencode, RefSeq, and Ensembl as reference. An FPKM > 1.5 was required in either SC or control cells. (D) Differentially expressed protein-coding, noncoding, pseudogene, and unannotated novel transcripts in SS.
The majority of transcripts detected in both malignant and nonmalignant T cells (72%) corresponded to annotated protein-coding genes (Figure 1C). Fifteen percent of detected transcripts were unannotated or uncharacterized in the UCSC, Ensembl, RefSeq, and Gencode gene databases and were designated as novel transcripts. Annotated noncoding genes and pseudogenes comprised the remaining 13% of expressed transcripts. These proportions are similar to those obtained recently by RNA-Seq in prostate tissues.2
We next identified aberrantly expressed transcripts in SS by selecting significantly changed annotated and novel genes in SCs compared with control lymphocytes. Annotated protein-coding (57%) and noncoding (24%) transcripts formed the majority of differentially expressed transcripts (Figure 1D), followed by novel transcripts (11%) and pseudogenes (8%). These differentially expressed transcripts provided candidates for further analysis.
Cancer-associated gene expression profile in SS
To assess protein-coding gene expression in SS, we assigned sequencing reads to coding transcripts using RefSeq annotations. Reads mapped to 24 888 protein-coding genes (of 32 668) and 19 550 (78.6%) were expressed in all 3 patients. Genes (n = 2989) were differentially expressed in all 3 patients; of these, 525 were commonly up-regulated and 519 were commonly down-regulated (Figure 2A). This shared signature of 1044 genes included several entities previously associated with SS, including up-regulation of TNFSF11 (RANKL), PTHLH, EPHA4, ZNF331, DDX41, KCNN4, ITGB1, CNIH4, and CD52 and down-regulation of APBA2, STAT4, NEDD4L, MXI1, TGFBR2, BCL2L11, SATB1, SP140, and RPS2.30–33
RNA-Seq in SS identities dysregulated cancer pathways and SC-associated IncRNAs. (A) Heat map of differentially expressed protein-coding genes. The expression of 2989 genes changed significantly in SC compared to patient-matched control cells (P < .05). Of these, 525 were commonly up-regulated and 519 were commonly down-regulated. (B) Significantly dysregulated pathways in SS. Canonical pathway analysis was performed with IPA using genes that changed significantly in all 3 patients. The significance of the association between this dataset and the canonical pathway shown was measured by Fisher's exact test. (C) Gene set enrichment analysis. GSEA plots depict concordance between significantly changed genes in SS and the molecular mechanisms of cancer canonical pathway from IPA (left), phosphatidylinositol signaling pathway (middle), and TGFβ signaling (right). NES, normalized enrichment score; FDR q-val, false discovery rate q-value (the probability that a gene set with a given NES represents a false-positive finding). (D) Heat map of differentially expressed IncRNAs. 35 were commonly up-regulated and 50 were commonly down-regulated. (E) Graphical representation of the bioinfomatic filters used to identify SC-associated IncRNAs. Expressed RefSeq transcripts annotated as noncoding were compiled with non-redundant, noncoding Gencode transcripts to assemble a merged SS noncoding transcriptome and the filters noted were applied. (F) Genomic locus of AL513543.1, a noncoding transcript that is down-regulated in the SC of all 3 patients. Histograms have been normalized to account for differences in the number of reads per library. (G) Relative expression of 21 SC-associated IncRNAs across RNA-Seq human tissue datasets normalized to average RPKM in polyclonal CD4+ T-cells. Gray boxes indicate no detectable expression. Name color indicates directionality of expression change in SC (red = increased, blue = decreased).
RNA-Seq in SS identities dysregulated cancer pathways and SC-associated IncRNAs. (A) Heat map of differentially expressed protein-coding genes. The expression of 2989 genes changed significantly in SC compared to patient-matched control cells (P < .05). Of these, 525 were commonly up-regulated and 519 were commonly down-regulated. (B) Significantly dysregulated pathways in SS. Canonical pathway analysis was performed with IPA using genes that changed significantly in all 3 patients. The significance of the association between this dataset and the canonical pathway shown was measured by Fisher's exact test. (C) Gene set enrichment analysis. GSEA plots depict concordance between significantly changed genes in SS and the molecular mechanisms of cancer canonical pathway from IPA (left), phosphatidylinositol signaling pathway (middle), and TGFβ signaling (right). NES, normalized enrichment score; FDR q-val, false discovery rate q-value (the probability that a gene set with a given NES represents a false-positive finding). (D) Heat map of differentially expressed IncRNAs. 35 were commonly up-regulated and 50 were commonly down-regulated. (E) Graphical representation of the bioinfomatic filters used to identify SC-associated IncRNAs. Expressed RefSeq transcripts annotated as noncoding were compiled with non-redundant, noncoding Gencode transcripts to assemble a merged SS noncoding transcriptome and the filters noted were applied. (F) Genomic locus of AL513543.1, a noncoding transcript that is down-regulated in the SC of all 3 patients. Histograms have been normalized to account for differences in the number of reads per library. (G) Relative expression of 21 SC-associated IncRNAs across RNA-Seq human tissue datasets normalized to average RPKM in polyclonal CD4+ T-cells. Gray boxes indicate no detectable expression. Name color indicates directionality of expression change in SC (red = increased, blue = decreased).
To look for evidence of dysregulation of specific signaling pathways in SS, we performed canonical pathway analysis with IPA (Ingenuity Systems; www.ingenuity.com) using genes that are differentially expressed in all 3 patients (Figure 2B). The most significantly affected pathways are molecular mechanisms of cancer (P < 2.3 × 10−12), followed by PI3K/Akt signaling (P < 2.5 × 10−12), and T-cell receptor signaling (P < 6.3 × 10−11). We also performed pathway analysis on genes that were expressed > 2-fold higher in SCs than in controls in all 3 patients to identify the pathways that are most robustly increased in SS. We found significant enrichment for genes in the NF-κB signaling cascade (P < 2.0 × 10−4), consistent with a prior report that NF-κB is constitutively active in CTCL cells.9,34 As a complement to IPA, we next performed GSEA with the use of annotated gene sets from the Molecular Signatures Database25 to determine concordance between significantly changed genes in SS and known gene signatures (Figure 2C). This analysis confirmed that the SS gene expression signature is enriched for genes with established importance in cancer as well as phosphatidylinositol and TGFβ signaling.
Differential expression of Sézary cell–associated lncRNAs
To identify differentially expressed lncRNAs in SS, we mapped sequencing reads to annotated noncoding transcripts in the RefSeq and Gencode databases. Reads mapped to 5099 noncoding transcripts (of 6629), 3711 (73%) of which are expressed in all 3 patients. The levels of 795 annotated noncoding transcripts are significantly (P < .05) or > 2-fold changed in all 3 patients. To enrich for lncRNAs, we removed transcripts from further analysis if they were < 200 bp in length, annotated pseudogenes, noncoding transcripts of protein-coding genes, or contained > 80% overlap with a known protein-coding transcript. We identified 258 transcripts that passed these filters; of these, 35 were commonly up-regulated and 50 were commonly down-regulated in SCs (Figure 2D).
To further refine the candidate pool, we required at least a 2-fold change in all 3 patients to enrich for robustly changed lncRNAs. We then reconstructed each of the remaining candidates with Trinity as well as Scripture and removed them from further analysis if the reassembled transcript was incongruent with the existing RefSeq or Gencode annotation. This workflow identified 21 congruently reconstructed lncRNAs with > 2-fold differential expression in all 3 patients (Figure 2E-F). Expression of these 21 SC-associated lncRNAs was generally detectable in a variety of human tissues (Figure 2G). These loci are largely uncharacterized and thus represent candidates for further study in SS.
Viral transcripts are not detected in SS cells
A viral cause of CTCL has been postulated, but focused searches for human T-lymphotropic virus 1 and human herpes viruses in CTCL have been inconclusive.35 To determine whether viral transcripts are detectable in the RNA-Seq data from the 3 patients with SS, we surveyed reads from SCs and control lymphocytes for uniquely viral sequences with an intersection-based approach that we validated with prostate adenocarcinoma cells infected with human papilloma virus-18 (NCBI Sequence Read Archive, SRR073726).26 Two sequence contigs with viral homology to human immunodeficiency virus and woodchuck hepatitis virus were generated, although subsequent PCR verification and Sanger sequencing confirmed both candidates to be laboratory contaminants (data not shown). Importantly, no sequences with homology to human T-lymphotropic virus 1 or other human viruses were identified in either SCs or control lymphocytes. These data therefore do not identify a potential viral etiology of SS.
Recurrent gene fusions are not detected in SS cells
Whole transcriptome sequencing data can be used to detect expressed gene fusions and has led to the discovery of recurrent gene fusions in several cancers.36,37 Prior studies have pointed to significant genomic instability in SCs; however, recurrent gene fusions have not been identified in CTCL or the majority of T-cell lymphomas in general.38,39 To determine whether any gene fusions are expressed in SCs or patient-matched normal control cells, we analyzed our paired-end sequencing reads by 2 orthogonal approaches using the deFuse and ChimeraScan algorithms.27,28 The former predicted one putative recurrent gene fusion event that did not validate by PCR amplification in the corresponding cDNA, whereas the latter did not nominate any candidate chimeric transcripts (data not shown). Importantly, the depth of coverage in our RNA-Seq data was comparable with that of recent work identifying CIITA rearrangements in Hodgkin lymphoma cell lines with the use of deFuse.37 Although we did not find recurrent gene fusions in SS, we cannot rule out translocations that would not yield fusion transcripts, such as those linking heterologous enhancers to an oncogene.
Novel differentially expressed transcripts in SS
To detect previously unidentified transcripts in SS that would have evaded detection by conventional DNA microarray approaches, de novo transcriptome assembly was performed with Scripture, and all annotated transcripts were removed.13 Coding potential scores were calculated for these novel transcripts and compared to the scores of known protein-coding and noncoding transcripts.40 Interestingly, novel transcripts as a group are generally predicted to be noncoding, a finding that may speak to the relatively recent emergence of ncRNA discovery efforts (supplemental Figure 1).
To determine whether these novel transcripts are differentially expressed in SS, we identified candidates that are concordantly changed > 2-fold in SCs compared to control T cells in all 3 patients. For this analysis, each transcript was required to contain > 1 exon, to be > 200 bp in length, and to have at least moderate abundance in either SCs or control cells. These steps were aimed at eliminating lowly expressed, single-exon, unreliable fragments generated by RNA-Seq. We identified 412 novel transcripts that met these criteria and eliminated entities that mapped to multiple genomic loci by BLAST analysis. Because transcript reconstruction algorithms frequently overreport isoforms, we further focused our analysis by manually curating the histograms of the read coverage of each transcript to collapse the novel pool into distinct genomic loci. This workflow resulted in 38 candidate novel transcripts that were differentially expressed in SS.
These 38 candidates were then independently reconstructed with Trinity and removed from further analysis if the resulting prediction diverged from the reconstruction generated by Scripture. Recent work comparing the congruence between lncRNA transcripts reconstructed by 2 assemblers shows that nearly half are identified by only 1 algorithm, consistent with discrepancies seen in the reconstruction of low-abundance protein-coding transcripts.21 As such, the use of Trinity as an orthogonal assembler to complement transcript predictions made by Scripture was designed to reduce computational artifacts generated by either assembler alone.
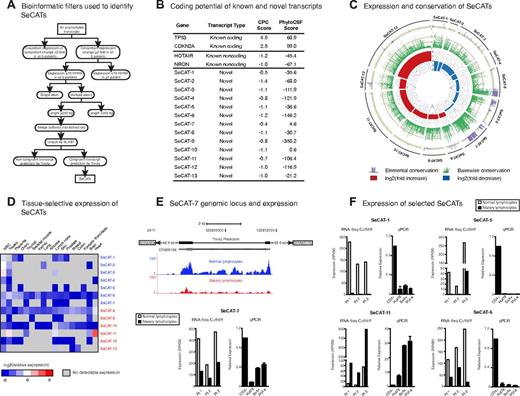
This analysis identified 13 novel Sézary cell–associated transcripts (SeCATs) that were congruently reconstructed and differentially expressed in all 3 patients with SS (Figure 3A; supplemental Table 3). We translated each SeCAT in all 6 reading frames and confirmed that none contained homology to any of the 31 912 known protein family domains documented in the Pfam database.41 To further enrich for bona fide noncoding transcripts in this group, we complemented our initial coding potential analysis with a second, independent assessment of coding capacity using PhyloCSF,24 an algorithm that uses sequence substitutions within a multispecies nucleotide alignment to determine whether evolutionary pressure exists to preserve an open reading frame (Figure 3B).21 Ten SeCATS are > 1000 times more likely to be noncoding than coding by PhyloCSF.
RNA-Seq in SS identifies novel Sézary cell-associated transcripts. (A) Graphical representation of the bioinformatic filters used to identify SeCATs. Previously unannotated transcripts were filtered by the pipeline shown to identify 13 candidates that met the criteria noted. (B) Coding potential of known and novel transcripts. Coding potential was measured using the CPC as well as PhyloCFS for 13 novel transcripts. The scores for selected canonical annotated protein-coding and noncoding transcripts are also shown for reference. (C) Expression and conservation of SeCATs. Elemental phastCons conservation across a given transcript (purple; interval between 0 and 0.2 is shaded in yellow, scores > 0.2 are considered conserved), basewise phyloP conservation (green; orange lines = S.D. > average genomic phyloP score), and expression log2 fold change (red = increase, blue = decrease) for 13 SeCATs. Each line on the outside of the circle depicts a SeCAT transcript and is drawn to scale; for reference, the transcript length of SeCAT-10 is 1.1 kb. (D) Relative expression of 13 SeCATs across RNA-Seq human tissue datasets normalized to average RPKM in polyclonal CD4+ T-cells. Gray boxes indicate no detectable expression. Name color indicates directionality of expression change in SC (red = increased, blue = decreased). (E) Genomic locus of SeCAT-7, a novel transcript that is down-regulated in the SC of all 3 patients. Histograms for 1 representative patient have been normalized to account for differences in the number or reads per library (top). The transcript structure predicted by Trinity as well as an unstudied cDNA clone with predicted coding potential and closest flanking genes are shown. Expression levels of SeCAT-7 in normal lymphocytes and SC in each of the 3 patients sequenced. Expression is shown as RPKM and calculated by Scripture (bottom left). Expression of SeCAT-7 by qPCR in CTCL cell lines (bottom right). The average value of CD4+ T-cells from 3 normal donors was used as control. (F) Expression levels of SeCAT-1, 5, 6, and 11 in normal lymphocytes and SC in each of the 3 patients sequenced is shown as RPKM and calculated by Scripture. Expression of SeCAT-1, 5, 6, and 11 is also demonstrated by qPCR in CTCL cell lines. The average value of CD4+ T-cells from 3 normal donors was used as control.
RNA-Seq in SS identifies novel Sézary cell-associated transcripts. (A) Graphical representation of the bioinformatic filters used to identify SeCATs. Previously unannotated transcripts were filtered by the pipeline shown to identify 13 candidates that met the criteria noted. (B) Coding potential of known and novel transcripts. Coding potential was measured using the CPC as well as PhyloCFS for 13 novel transcripts. The scores for selected canonical annotated protein-coding and noncoding transcripts are also shown for reference. (C) Expression and conservation of SeCATs. Elemental phastCons conservation across a given transcript (purple; interval between 0 and 0.2 is shaded in yellow, scores > 0.2 are considered conserved), basewise phyloP conservation (green; orange lines = S.D. > average genomic phyloP score), and expression log2 fold change (red = increase, blue = decrease) for 13 SeCATs. Each line on the outside of the circle depicts a SeCAT transcript and is drawn to scale; for reference, the transcript length of SeCAT-10 is 1.1 kb. (D) Relative expression of 13 SeCATs across RNA-Seq human tissue datasets normalized to average RPKM in polyclonal CD4+ T-cells. Gray boxes indicate no detectable expression. Name color indicates directionality of expression change in SC (red = increased, blue = decreased). (E) Genomic locus of SeCAT-7, a novel transcript that is down-regulated in the SC of all 3 patients. Histograms for 1 representative patient have been normalized to account for differences in the number or reads per library (top). The transcript structure predicted by Trinity as well as an unstudied cDNA clone with predicted coding potential and closest flanking genes are shown. Expression levels of SeCAT-7 in normal lymphocytes and SC in each of the 3 patients sequenced. Expression is shown as RPKM and calculated by Scripture (bottom left). Expression of SeCAT-7 by qPCR in CTCL cell lines (bottom right). The average value of CD4+ T-cells from 3 normal donors was used as control. (F) Expression levels of SeCAT-1, 5, 6, and 11 in normal lymphocytes and SC in each of the 3 patients sequenced is shown as RPKM and calculated by Scripture. Expression of SeCAT-1, 5, 6, and 11 is also demonstrated by qPCR in CTCL cell lines. The average value of CD4+ T-cells from 3 normal donors was used as control.
To further characterize these 13 novel transcripts, we analyzed their basewise conservation by phyloP and noted that they display increased conservation across their exons compared with the genome average (Figure 3C). SeCAT conservation scores were comparable with the scores of known noncoding transcripts (supplemental Figure 2). We then used phastCons to assess for conserved elements within SeCAT mRNAs and identified highly conserved regions in multiple transcripts, consistent with potential functional importance (Figure 3C).42 In contrast to SC-associated lncRNAs that are generally detectable in a wide variety of tissues, SeCATs are largely composed of CD4+ T cell– or SC-specific transcripts and contain only a few entities with broad tissue expression (Figure 3D).
We next sought to study whether the differential SeCAT expression observed in freshly purified SCs and normal T cells from blood also occurred in CTCL cell lines by studying a subset of robustly expressed SeCATs in more depth. We compared the expression of SeCAT-1, -5, -6, -7, -9, and -11 in 3 CTCL cell lines (MyLa, SeAx, and Hut-78). RNA levels in these lines were compared with pooled CD4+ T cells from 3 healthy donors. Concordance in the directionality of expression level changes was seen for all SeCATs studied, with the exception of SeCAT-9 (Figure 3E-F; supplemental Figure 3A-B). Although SeCAT-9 is up-regulated in the SCs of all 3 patients with SS, it is expressed at far lower levels in CTCL cell lines than in normal CD4+ T cells (supplemental Figure 3B). Inspection of SeCAT-9 sequencing histograms confirmed up-regulation of this 3 exon transcript in SCs (supplemental Figure 3A), as did qRT-PCR validation in 2 additional freshly sorted SS patient SCs (patients 4 and 5) versus their matched CD4+ T cells as controls (supplemental Figure 3B). SeCAT-9 is therefore up-regulated in 5 of 5 patient SCs but down-regulated in all 3 widely used CTCL cell lines, underscoring the potential lack of concordance between expression of a subset of transcripts in these 2 different contexts. Thus, the altered expression of most, but not all, SeCATs is recapitulated in CTCL cell lines.
Differential expression of Sézary cell–associated lncRNAs and SeCATs in MF
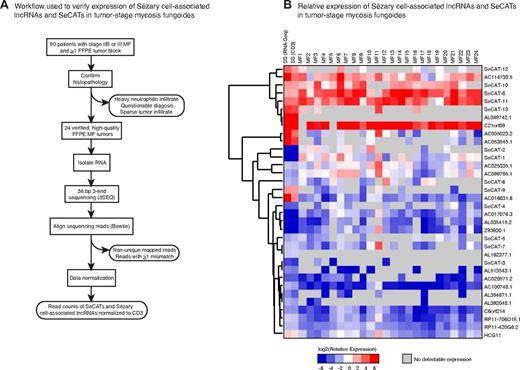
To determine whether differential expression of SC-associated lncRNAs and SeCATs are also present in MF, we performed high-throughput RNA sequencing to target the 3′ end of polyadenylated transcripts (3SEQ) on 24 archival MF tumors (Figure 4A). This approach allows quantification of transcript abundance in FFPE tissue despite partial RNA degradation.9 We obtained an average of 42 million total reads for each sample and an average of 13 million unique mapped reads for each sample. This depth of coverage reflects a 2.5-fold enrichment of unique mapped base pairs compared with a previous report that used FFPE samples.9
Validation of SC-associated IncRNAs and SeCATs in MF. (A) Schematic of experimental design. The 3' ends of polyadenylated transcripts from 24 high-quality FFPE MF tumor specimens were sequenced by 3SEQ. Sequence reads were aligned to the genome and filtered by the pipeline shown. (B) Relative expression of 21 SC-associated IncRNAs and 13 SeCATs across 24 MF tumors normalized to average CD3 read count and then compared to its average expression relative to CD3 in polyclonal CD4+ T-cells from SS patients; average expression of SC-associated IncRNAs and SeCATs in SS normalized to their expression in patient-matched polyclonal CD4+ T-cells (RNA-Seq) as well as to CD3 is also shown. Gray boxes indicate no detectable expression.
Validation of SC-associated IncRNAs and SeCATs in MF. (A) Schematic of experimental design. The 3' ends of polyadenylated transcripts from 24 high-quality FFPE MF tumor specimens were sequenced by 3SEQ. Sequence reads were aligned to the genome and filtered by the pipeline shown. (B) Relative expression of 21 SC-associated IncRNAs and 13 SeCATs across 24 MF tumors normalized to average CD3 read count and then compared to its average expression relative to CD3 in polyclonal CD4+ T-cells from SS patients; average expression of SC-associated IncRNAs and SeCATs in SS normalized to their expression in patient-matched polyclonal CD4+ T-cells (RNA-Seq) as well as to CD3 is also shown. Gray boxes indicate no detectable expression.
Although a pure population of patient-matched control cells was obtained for each SS sample studied, procurement of appropriate control tissue for MF specimens was complicated by variations in tumor heterogeneity across samples. To quantify the expression of SC-associated lncRNAs and SeCATs in MF tumors, we normalized the read count of each transcript to the averaged CD3 read count in that sample (Figure 4B; supplemental Table 4). Expression of CD3 is limited to T cells and was not significantly changed between SCs and polyclonal CD4+ T cells. The expression of each transcript of interest relative to CD3 was then compared with its average expression relative to CD3 in polyclonal CD4+ T cells from patients with SS. This approach correctly identified the directionality of expression changes for all SC-associated lncRNAs and SeCATs in SS, providing proof-of-principle for this method of normalization and justifying its application to our MF data. Detectable expression of SC-associated lncRNAs and SeCATs was generally observed in MF tumors, with only 5 of 34 (15%) of these transcripts detected in < 6 MF samples. Concordance in the directionality of expression level changes was seen for 14 of 21 (67%) SC-associated lncRNAs and 8 of 13 (62%) SeCATs (Figure 4B). RNA degradation, transcriptional differences between MF and SS, and the relatively small discovery set of 3 patients with SS probably account for the nonconcordantly expressed transcripts. Interestingly, most SC-associated lncRNA and SeCAT expression patterns are recapitulated in the set of 24 MF tumors, supporting our findings from RNA-Seq analysis of SS and highlighting possible transcriptional similarities between SS and MF.
Discussion
Recent advances in high-throughput sequencing provide the opportunity to characterize cancer genomes at unprecedented depth and identify key oncogenic pathways as well as putative therapeutic targets. We report whole transcriptome analysis of 3 patients with SS and compare gene expression in freshly isolated SCs with patient-matched polyclonal CD4+ T cells. The use of purified SCs and their comparison to autologous polyclonal CD4+ T cells was designed to obtain as pure a cell population as possible while minimizing detection of less relevant differences in RNA expression due to environmental factors, genetic heterogeneity, and long-term adaptation to cell culture characteristic of cancer cell lines. To minimize detection of transcriptional changes due to treatment effects, the 3 patients selected each received differing therapies at the time of sample collection and gene expression changes were calculated relative to each patient's polyclonal CD4+ population; however, we cannot rule out changes that resulted from selective targeting of SCs but not nonmalignant CD4+ T cells. The data identify transcriptional mis-regulation of PI3K/Akt, TGFβ, NF-κB, and T-cell receptor signaling pathways in SS, consistent with previous reports.31,34 The data also identify 13 previously unreported transcripts that we have termed SeCATs as well as 21 annotated SC-associated lncRNAs that are differentially expressed in SCs versus normal CD4+ T cells and in the majority of an independent set of 24 MF tumors.
Analysis of differentially expressed protein-coding genes produced a core signature of 1044 genes that are concordantly changed in each of the 3 patients with SS sequenced. Although different groups have previously described expression signatures in SS, the heterogeneity of experimental conditions across studies has obscured a clear consensus. To our knowledge, only 1 prior study that described a single patient compared SCs with patient-matched T cells using DNA microarray technology.32 Of the 44 differentially expressed genes in that study, 13 (30%) were significantly changed in all 3 of the patients with SS studied here (supplemental Table 5), highlighting the power of sorting cells by this approach. The remaining divergence between datasets may relate to the genetic diversity or to the use of different experimental platforms.
Our analysis also found increased expression of several genes that encode transmembrane proteins in all 3 patients (supplemental Table 6). Presenilin-1 is part of the γ-secretase complex that activates Notch1, a potential therapeutic target in CTCL and an important oncogenic pathway in T-cell acute lymphoblastic leukemia.43 KCNN4 is a calcium-regulated potassium channel implicated in T-cell activation and proliferation; inhibition of this channel by TRAM-34 decreases inflammation in murine models of encephalomyelitis and colitis,44 raising the possibility that it may also reduce SC proliferation. We further note overexpression of PDCD1 in SCs, consistent with previous reports.45 Given their accessibility to therapeutic antibodies, the transmembrane proteins up-regulated in SS comprise attractive targets for future study.
Prior studies have investigated a potential link between CTCL and viral infections.35 The absence of viral transcripts in our dataset argues against a viral etiology in SS, although transcriptionally silent viral integration events would escape detection by RNA-Seq. Further, translocations that deregulate oncogene expression by altering its proximity to enhancers, such as the MYC-IGH rearrangement,39 could escape detection by RNA-Seq yet might be detected by whole genome sequencing. We also note that recurrent fusion genes occur in a minority of mature T-cell neoplasms, and chromosomal translocations that involve the TCR locus in CTCL have not been found.46
LncRNAs have not previously been reported in association with SS or MF, but they have been described in other human cancers and exert functional effects on diverse cellular processes.2–4 To our knowledge, no exhaustive search for cancer-related lncRNAs has been conducted in T cells. We surveyed our data for differentially expressed annotated lncRNAs and identified 21 that change concordantly and robustly in the 3 SS individuals sequenced. In contrast to SeCATs that display high tissue selectivity, expression of SC-associated lncRNAs was detected in a broad spectrum of normal human tissues. The more ubiquitous expression of these previously annotated lncRNAs probably relates to their identification in nontransformed tissues containing few T cells and suggests that these transcripts may serve broader biologic functions.
These data also identify 13 novel transcripts with differential expression in SS. The SeCATs are largely predicted to be noncoding, although we cannot rule out the possibility that they may share features with short, polycistronic ribosome-associated RNAs or encode small peptides similar to those that regulate Drosophila embryogenesis.47,48 Recent large-scale efforts to characterize lncRNAs have identified a number of common features that help distinguish them from protein-coding genes, including higher tissue specificity, less evolutionary conservation, and lower overall expression.21 We examined SeCAT coordinates in publicly available RNA-Seq datasets from a variety of normal human tissues and observed that SeCATs display high tissue selectivity. They also demonstrate modest conservation comparable with noncoding transcripts, and several contain substantial elemental conservation that may suggest involvement in posttranscriptional regulation or RNA secondary structure. Although we focused on higher abundance candidates because they are more experimentally tractable, we do observe lower expression of novel transcripts in general compared to protein-coding genes. These data indicate that SeCATs contain features of lncRNAs.
Among the 3 SeCATs not designated by PhyloCSF as highly likely to be noncoding, SeCAT-7 had the highest positive score (Figure 3B,E), profound conservation at the nucleotide level, and an uncharacterized cDNA clone with a txCdsPredict score of 975 (scores > 800 have a 90% chance of being protein coding).16 Sequence analysis identified a putative 408-bp open reading frame (ORF) that blasts to a hypothetical protein in nonhuman primates (E value < 1 × 10−72; supplemental Figure 4A), as well as a confirmed protein in other placental mammals, including Mus musculus and Bos taurus (E value < 7 × 10−57) and a predicted protein in nonmammals including Oreochromis niloticus (tilapia) and Tetraodon nigroviridis (pufferfish; E value < 3 × 10−19). We also noted a putative 811-bp ORF that begins with a noncanonical start codon and blasts to a hypothetical protein in Pongo abelii (orangutan) and Nomascus leucogenys (gibbon; E value < 3 × 10−61; supplemental Figure 4B). Analysis of genomic sequence in the nearest nonhuman primate Pan troglodytes verified this preserved open reading frame, although RNA-Seq has not been widely applied to nonhuman primate tissues, and no transcript has yet been described in chimpanzees. We postulate that the smaller ORF may be coding, but note that noncanonical and AUG start codons can coexist at the same genic locus and may encode alternate isoforms with distinct functions as well as regulatory roles.49 Thus, SeCAT-7 may represent an uncharacterized protein-coding transcript.
Current estimates place the incidence of SS at 30 to 150 new cases per year in the United States, and in our experience < 50% of these patients have a detectable Vβ clone. These limitations, as well as the practical requirement that persons have sufficiently high counts of both normal and malignant cell populations to allow comparison of sorted T cells highly enriched for the malignant TCR clone with polyclonal T cells, complicate the prospective collection of a large validation set of patients with SS. We therefore sought to verify differential expression of SC-associated lncRNAs and SeCATs using archival specimens and performed 3SEQ on 24 MF tumors from individuals with stage IIB/III disease. Examination of the coordinates of SC-associated lncRNAs and SeCATs in the 3SEQ dataset revealed discernable expression in most of these samples, and the directionality of their relative enrichment in MF matched that seen in SS in > 60% of these transcripts. These findings confirm the existence of previously unidentified SeCATs in MF, suggest that SC-associated lncRNAs and SeCATs represent transcripts with differential expression, and provide orthogonal support for the bioinformatic filters used to distill these candidates from the initial RNA-Seq screen. Although studies have suggested that SS and MF may represent genetically distinct entities, these similarities in the differential expression of SC-associated lncRNAs and SeCATs in these 2 diseases raises the intriguing possibility of overlapping transcriptomic features. Future studies are needed to address the functional significance of SC-associated lncRNAs and SeCATs as well as to elucidate their mechanisms of action.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Veterans Affairs Office of Research and Development, the Haas Family Fund, and the National Institutes of Health [grants RO1AR49737 (P.A.K.), F32 CA142135 (C.S.L.), 5T32HL007970 (A.U.), and 5T32CA009302 (A.B.)].
National Institutes of Health
Authorship
Contribution: C.S.L., A.U., and P.A.K. designed the research; C.S.L., A.U., A.B., C.J.A., and A.M. performed experiments; R.A. performed flow cytometry; R.O.C. collected MF tumor blocks; M.L. prepared the 3SEQ sequencing libraries; U.S. reviewed the histopathology of MF specimens; C.S.L., A.U., A.B., K.Q., and D.E.W. performed bioinformatic analysis; C.S.L., A.U., A.B., and P.A.K. analyzed results; C.S.L. wrote the manuscript and made the figures; A.U., A.B., D.E.W., K.Q., W.-K.W., H.Y.C., M.K., Y.H.K., and P.A.K. provided experimental insight and revised the manuscript.
Conflict-of-interest disclosure: R.O.C. is a shareholder of Ingenuity Systems, Inc. The remaining authors declare no competing financial interests.
Correspondence: Paul A. Khavari, 269 Campus Dr, Rm 2145, Stanford, CA 94305; e-mail: khavari@stanford.edu.