Abstract
In patients with follicular lymphoma treated with single-agent rituximab, single nucleotide polymorphisms in the FCGR3A gene are known to influence response and progression-free survival. The prognostic role of FCGR3A and FCGR2A polymorphisms in patients with follicular lymphoma treated with rituximab and chemotherapy combination remains controversial and has not been evaluated in the context of rituximab maintenance. FCGR3A and FCGR2A single nucleotide polymorphisms were evaluated in, respectively, 460 and 455 patients treated in the PRIMA study to investigate whether these were associated with response rate and patient outcome after rituximab chemotherapy induction and 2-year rituximab maintenance. In this representative patient cohort, complete and unconfirmed complete responses after rituximab chemotherapy were observed in 65%, 67%, 66% (P = .86) and 60%, 72%, 66% (P = .21) of FCGR3A VV, VF, FF and FCGR2A HH, HR, RR carriers, respectively. After 2 years of rituximab maintenance (or observation), response rates did not differ among the different genotypes. Progression-free survival measured from either treatment initiation or randomization to observation or maintenance was not influenced by these polymorphisms. These data indicate that FCGR3A and FCGR2A polymorphisms do not influence response rate and outcome when rituximab is combined with chemotherapy or used as maintenance treatment. The PRIMA study is registered at www.clinicaltrials.gov as NCT00140582.
Introduction
The prognosis of patients with follicular lymphoma (FL) remains variable, and the natural history of this lymphoma is typified by multiple episodes of relapse.1 Despite a relatively long overall survival (OS), advanced stage FL generally remains an incurable disease. To more accurately predict a patient's prognosis, a variety of scores that are based on presenting clinical and simple biologic characteristics, such as the follicular lymphoma international prognostic index (FLIPI) score, have been proposed.2 The decision to initiate treatment is usually determined by the presence of a clinically significant tumor mass, hematopoietic compromise, or symptomatic disease.3
Recent biologic data have suggested that the host–tumor interaction may influence disease behavior, and assessment of specific cellular components within the tumor microenvironment may provide additional prognostic information.4 The outcome of patients could also be influenced by inherent host immunologic factors. For example, recent studies have shown that the prognosis of patients with FL was associated with germline polymorphisms in immune-response elements, such as the cytokine genes IL1RN, IL2, IL8, and IL12.5 The prognosis of patients with FL has dramatically improved with the introduction of anti-CD20 monoclonal antibodies (mAbs).6-9 The therapeutic activities of these mAbs may also be affected by patient biologic characteristics such as polymorphisms in FcG receptors (FCγR) genes.10 The affinity of the Fc portion of anti-CD20 mAb on FCγRs is modulated by specific single nucleotide polymorphisms (SNPs) in FCGR genes. For instance, the SNP rs396991 leads to the amino acid substitution of a valine (V) for a phenylalanine (F), the FCγRIIIA 158V having a higher affinity of human immunoglobulin G1 (hu-Ig G1) than FCγRIIIA F allele.11 Similarly, the polymorphism (rs1801274) in FCGR2A gene lead to FcγRIIA with either a histidine (H) or an arginine (R) at amino acid position 131, where the FCγRIIA 131H variant binds more strongly to hu-Ig G1.11 The possible clinical consequence is a difference in the quality and the duration of response to anti-CD20 mAbs. Some clinical studies have shown that patients homozygous for FCGR3A 158VV have an improved response to treatment with single-agent rituximab.12,13 However, the effect of this polymorphism on longer-term outcome12-14 as well as its consequences in the context of immunochemotherapy are more controversial.15,16 The biologic and clinical relevance of others SNPs such as FCGR2A in patients with FL also need to be clarified.12,13
The PRIMA study recently showed that patients with high tumor burden FL that responded to immunochemotherapy have improved progression-free survival (PFS) after 2 years of rituximab maintenance.17 Prespecified ancillary biologic objectives of the PRIMA study included clarification, in the context of a prospective trial, of the role of intrinsic FCGR polymorphisms in patients with FL treated with rituximab and chemotherapy (R-CT) and to determine their prognostic effect for patients treated by rituximab maintenance after R-CT. Our genetic studies focused on FCGR3A and FCGR2A polymorphisms in, respectively, 460 and 455 patients treated in the PRIMA study.
Methods
Study population
Peripheral-blood DNA samples were prospectively obtained from 460 patients with FL included in the open-label, international, multicenter randomized PRIMA study that enrolled a total of 1217 patients with untreated high tumor burden FL.17 Only patients with confirmed grade 1, 2, or 3a FL were included in the present biologic study. The FLIPI scores were assessed before treatment. Patients were first treated by 1 of the 3 protocol-specified standard immunochemotherapy regimens (induction phase). The 3 rituximab combinations used in the PRIMA study were CVP (cyclophosphamide 750 mg/m2 on day 1, vincristine 1.4 mg/m2 [capped at 2 mg] on day 1, prednisone 40 mg/m2 on days 1-5, repeated every 3 weeks for 8 cycles), CHOP (cyclophosphamide 750 mg/m2 on day 1, vincristine 1.4 mg/m2 [capped at 2 mg] on day 1, doxorubicin 50 mg/m2 on day 1, prednisone 100 mg on days 1-5, repeated every 3 weeks for 6 cycles), and FCM (fludarabine 25 mg/m2 on days 1-3, cyclophosphamide 200 mg/m2 on days 1-3, mitoxantrone 6 mg/m2 on day 1, repeated every 4 weeks for 6 cycles). Rituximab (375 mg/m2 at each infusion) was administered on day 1 of each chemotherapy course. Two additional rituximab infusions were administered to patients treated with CHOP (every 3 weeks after the last cycle) and FCM (2 weeks after the first and the fourth cycles) to ensure equivalent exposure to the antibody during induction for all patients. Response to this induction therapy was assessed 2-4 weeks after the last treatment course. After this induction phase, patients who obtained a complete response (CR), an unconfirmed CR (CRu), and a partial response (PR) were randomly assigned in a 1:1 ratio to observation or rituximab maintenance (12 infusions of 375 mg/m2 at 8-week intervals). Patients were evaluated clinically every 8 weeks during the 2-year maintenance phase and by computed tomographic scan every 6 months. Patients with bone marrow involvement at diagnosis underwent bone marrow evaluation at the end of the maintenance phase. Thereafter, a clinical evaluation and a computed tomographic scan were performed, respectively, every 3 and 6 months for a period of 3 years. This study was conducted in accordance with the Declaration of Helsinki. All patients signed a consent form for participation in specific studies of germline polymorphisms approved by the ethics committees of Lyon University Hospitals. The PRIMA study is registered with www.clinicaltrials.gov as NCT00140582.
Laboratory analysis
DNA was extracted from peripheral blood mononuclear leukocytes before any treatment. The 2 SNPs in FCGR3A and FCGR2A were named according to the SNP500 Cancer database and identified according to the ID numbers of the NCBI dbSNP database, rs396991 and rs1801274, respectively.18 The SNP in FCGR3A was analyzed with specific fluorescent dye–labeled (FAM and VIC) MGB probes (Applied Biosystems). Real-time PCR analysis was performed on an ABI Prism 7000 Sequence Detection System (Applied Biosystems) with the use of a total volume of 25 μL with 2 μL of DNA (5 ng/μL). The forward and reverse primer pairs and MGB probes were obtained from the SNP500 Cancer database. PCR conditions were as follows: 50°C for 2 minutes, 95°C for 10 minutes, and 35 cycles at 92°C for 15 seconds and 67°C for 60 seconds. FCGR2A genotyping varied slightly, because a complete commercially available assay (Applied Biosystems) that contained primers, probes, and TaqMan Genotyping Master Mix was used. The amplification program for FCGR2A consisted of 50°C for 2 minutes, 95°C for 10 minutes, and 40 cycles at 95°C for 15 seconds and 60°C for 60 seconds. All analyses were performed in duplicate.
Statistical analysis
Clinical characteristics, prognostic parameters, response to induction treatment, and outcome were compared between the 460 patients available for FCGR genotyping and all patients included in the PRIMA study to ensure that our study sample was representative of the general patient population. Response to induction phase was assessed according to Cheson criteria.19 Patients could be classified as having a CR, a CRu, a PR, or no response with stable or progressive disease. Response was also evaluated at the end of the 2-year rituximab maintenance phase with the use of similar response criteria. PFS and OS defined by international criteria could be evaluated from the date of registration before induction phase or from the time of randomization to rituximab maintenance or observation.19 The correlation among FCGR genotypes and initial characteristics, response to treatment, and outcome were assessed. A χ2 test was used to examine associations among genotypes and patient characteristics, treatment response, and treatment toxicities. Time-to-event parameters were estimated by the Kaplan-Meier product limit method and compared by log-rank test.
Results
Clinical results
The clinical characteristics of the 460 patients with FL of the FCGR cohort in this ancillary study were comparable with those of the whole PRIMA cohort (1193 patients with complete data who received induction treatment), except for the more-frequent presence of B symptoms in patients of the PRIMA study (33% vs 27%; P = .02; supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Most of the 460 analyzed patients received induction treatment with rituximab and CHOP (R-CHOP; n = 374; 81%). Other patients received rituximab and CVP (R-CVP; n = 84; 18%), or rituximab and FCM (R-FCM; n = 2; < 1%). A slightly different treatment distribution was reported in the PRIMA study: R-CHOP with n = 881 (74%), R-CVP with n = 268 (22%), and R-FCM with n = 44 (4%). Evaluation of response after induction therapy in the 460 analyzed patients of the FCGR study showed that 159 patients (35%) were in CR, 147 in CRu (32%), and 128 in PR (28%), and 21 patients (5%) had stable or progressive disease, but 5 patients (1%) could not be evaluated. Among the 1019 patients who were randomly assigned,17 after induction treatment, 398 had been analyzed for FCGR study, 196 patients in the observation arm and 202 into the maintenance arm; 1 patient in the maintenance arm died immediately after random assignment. We evaluated the primary end point (PFS in randomly assigned patients) of the PRIMA study in this subpopulation. With a median follow-up of 36 months, the 3-year PFS rate was 79.2% (95% CI, 72.6%-84.4%) in the rituximab maintenance arm and 56.2% (95% CI, 48.8%-63.0%) in the observation arm (P < .0001), thereby confirming the results of the PRIMA study (supplemental Figure 1).
Genotype frequencies of FCGR3A and FCGR2A alleles
FCGR3A was tested in all 460 patients; the frequency of the VV, VF, and FF alleles was 15%, 47%, and 38%, respectively. FCGR2A was tested in 450 patients; the distribution of the HH, HR, and RR alleles was 27%, 50%, and 23%, respectively. These polymorphism distributions were consistent with Hardy-Weinberg equilibrium. Because the PRIMA study is an international project, 78 genomic DNA samples were obtained from Australian and 382 from European (France and Belgium) patients. The allele distribution was not different among patients from different continents. In accordance with French law, no mention of race or ethnicity was made. Comparison of patient characteristics according to FCGR3A and FCGR2A allele status showed no difference between disease characteristics and each SNP. Similarly, the distribution of alleles for these 2 SNPs was similar among patients treated by R-CHOP, R-CVP, and R-FCM (Table 1).
Correlation between FCGR3A and FCGR2A genotypes and clinical characteristics
| . | FCGR3A . | FCGR2A . | ||||||
|---|---|---|---|---|---|---|---|---|
| VV, n (%) . | VF, n (%) . | FF, n (%) . | P . | HH, n (%) . | HR, n (%) . | RR, n (%) . | P . | |
| No. of patients | 68 (15) | 215 (47) | 177 (38) | * | 122 (27) | 226 (50) | 102 (23) | * |
| Europe | 53 (14) | 181 (47) | 148 (39) | — | 100 (27) | 184 (49) | 89 (24) | — |
| Australia | 15 (19) | 34 (44) | 29 (37) | — | 22 (29) | 42 (55) | 13 (17) | — |
| Male sex | 38 (56) | 120 (56) | 96 (54) | .94 | 62 (52) | 129 (57) | 57 (56) | .61 |
| Age > 60 y | 26 (38) | 90 (42) | 63 (36) | .44 | 43 (35) | 95 (42) | 38 (37) | .42 |
| Ann Arbor stage III/IV | 63 (93) | 195 (91) | 161 (91) | .88 | 115 (94) | 203 (90) | 92 (90) | .35 |
| ECOG PS ≥ 1 | 22 (32) | 69 (32) | 56 (31) | .91 | 31 (25) | 79 (35) | 34 (33) | .43 |
| B symptoms present | 16 (24) | 60 (28) | 47 (27) | .77 | 32 (26) | 60 (27) | 30 (29) | .83 |
| BM involvement | 38 (58) | 113 (53) | 101 (58) | .70 | 74 (62) | 122 (55) | 52 (51) | .58 |
| Elevated LDH | 16 (24) | 67 (31) | 65 (37) | .13 | 35 (29) | 74 (33) | 35 (29) | .71 |
| Hemoglobin level < 12 g/dL | 18 (26) | 39 (18) | 25 (14) | .07 | 22 (18) | 40 (18) | 17 (17) | .96 |
| β2-microglobulin ≥ 3 mg/L | 18 (28) | 52 (26) | 49 (30) | .69 | 26 (22) | 58 (28) | 31 (33) | .23 |
| FLIPI score | ||||||||
| 0-1 risk factors | 17 (25) | 51 (24) | 39 (22) | .64 | 26 (21) | 52 (23) | 27 (26) | .85 |
| 2 risk factors | 20 (29) | 71 (33) | 69 (39) | 47 (39) | 78 (35) | 33 (32) | ||
| 3-5 risk factors | 31 (46) | 92 (43) | 69 (39) | 49 (40) | 95 (42) | 42 (41) | ||
| Induction regimen | ||||||||
| R-CHOP | 54 (79) | 175 (81) | 145 (82) | .55 | 97 (80) | 181 (80) | 87 (85) | .23 |
| R-CVP | 13 (19) | 40 (19) | 31 (18) | 23 (19) | 45 (20) | 15 (15) | ||
| R-FCM | 1 (1) | 0 (0) | 1 (1) | 2 (2) | 0 (0) | 0 (0) | ||
| . | FCGR3A . | FCGR2A . | ||||||
|---|---|---|---|---|---|---|---|---|
| VV, n (%) . | VF, n (%) . | FF, n (%) . | P . | HH, n (%) . | HR, n (%) . | RR, n (%) . | P . | |
| No. of patients | 68 (15) | 215 (47) | 177 (38) | * | 122 (27) | 226 (50) | 102 (23) | * |
| Europe | 53 (14) | 181 (47) | 148 (39) | — | 100 (27) | 184 (49) | 89 (24) | — |
| Australia | 15 (19) | 34 (44) | 29 (37) | — | 22 (29) | 42 (55) | 13 (17) | — |
| Male sex | 38 (56) | 120 (56) | 96 (54) | .94 | 62 (52) | 129 (57) | 57 (56) | .61 |
| Age > 60 y | 26 (38) | 90 (42) | 63 (36) | .44 | 43 (35) | 95 (42) | 38 (37) | .42 |
| Ann Arbor stage III/IV | 63 (93) | 195 (91) | 161 (91) | .88 | 115 (94) | 203 (90) | 92 (90) | .35 |
| ECOG PS ≥ 1 | 22 (32) | 69 (32) | 56 (31) | .91 | 31 (25) | 79 (35) | 34 (33) | .43 |
| B symptoms present | 16 (24) | 60 (28) | 47 (27) | .77 | 32 (26) | 60 (27) | 30 (29) | .83 |
| BM involvement | 38 (58) | 113 (53) | 101 (58) | .70 | 74 (62) | 122 (55) | 52 (51) | .58 |
| Elevated LDH | 16 (24) | 67 (31) | 65 (37) | .13 | 35 (29) | 74 (33) | 35 (29) | .71 |
| Hemoglobin level < 12 g/dL | 18 (26) | 39 (18) | 25 (14) | .07 | 22 (18) | 40 (18) | 17 (17) | .96 |
| β2-microglobulin ≥ 3 mg/L | 18 (28) | 52 (26) | 49 (30) | .69 | 26 (22) | 58 (28) | 31 (33) | .23 |
| FLIPI score | ||||||||
| 0-1 risk factors | 17 (25) | 51 (24) | 39 (22) | .64 | 26 (21) | 52 (23) | 27 (26) | .85 |
| 2 risk factors | 20 (29) | 71 (33) | 69 (39) | 47 (39) | 78 (35) | 33 (32) | ||
| 3-5 risk factors | 31 (46) | 92 (43) | 69 (39) | 49 (40) | 95 (42) | 42 (41) | ||
| Induction regimen | ||||||||
| R-CHOP | 54 (79) | 175 (81) | 145 (82) | .55 | 97 (80) | 181 (80) | 87 (85) | .23 |
| R-CVP | 13 (19) | 40 (19) | 31 (18) | 23 (19) | 45 (20) | 15 (15) | ||
| R-FCM | 1 (1) | 0 (0) | 1 (1) | 2 (2) | 0 (0) | 0 (0) | ||
ECOG indicates Eastern Cooperative Oncology Group; PS, performance status; BM, bone marrow; LDH, lactate dehydrogenase; FLIPI, follicular lymphoma international prognostic index; R-CHOP, rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone; R-CVP, rituximab, cyclophosphamide, vincristine, and prednisone; and R-FCM, rituximab, fludarabine, cyclophosphamide, and mitoxantrone.
The 2 single nucleotide polymorphisms in FCGR3A and FCGR2A were tested for Hardy-Weinberg equilibrium.
Response to induction therapy according to FCGR3A and FCGR2A alleles
Among the 460 patients genotyped, respectively, 455 and 450 patients with FCGR3A and FCGR2A SNPs were assessable for response after induction therapy. The quality of response after immunochemotherapy was not influenced by the FCGR3A and FCGR2A allele status (Table 2). For FCGR3A polymorphism, CR/CRu was observed in 65% of patients with the VV allele, in 67% with the VF allele, and 66% with the FF allele. For FCGR2A polymorphism, CR/CRu was observed in 60% patients with the HH allele, 72% with the HR allele, and 66% with the RR allele. Results were similar when analyses were restricted to patients treated by R-CHOP induction therapy: 67%, 69%, and 71% of patients with VV, VF, FF FCGR3A alleles and 64%, 72%, and 69% of patients with HH, HR, RR FCGR2A alleles, respectively (Table 2).
Clinical response according to FCGR3A and FCGR2A genotypes
| . | FCGR3A . | FCGR2A . | ||||||
|---|---|---|---|---|---|---|---|---|
| VV, n (%) . | VF, n (%) . | FF, n (%) . | P . | HH, n (%) . | HR, n (%) . | RR, n (%) . | P . | |
| Response to induction | ||||||||
| Whole series | ||||||||
| CR | 23 (34) | 75 (35) | 61 (35) | .86 | 41 (34) | 75 (34) | 38 (38) | .21 |
| CRu | 21 (31) | 69 (32) | 57 (33) | 31 (26) | 85 (38) | 28 (28) | ||
| CR/CRu | 44 (65) | 144 (67) | 118 (68) | 72 (60) | 160 (72) | 66 (66) | ||
| PR | 21 (31) | 61 (29) | 46 (26) | 43 (36) | 53 (24) | 30 (30) | ||
| SD/PD | 3 (4) | 8 (4) | 10 (6) | 5 (4) | 11 (4) | 5 (5) | ||
| R-CHOP regimen | ||||||||
| CR/CRu | 36 (67) | 121 (69) | 103 (71) | .29 | 62 (64) | 131 (72) | 60 (69) | .55 |
| PR | 18 (33) | 48 (27) | 34 (23) | 32 (33) | 43 (24) | 23 (26) | ||
| SD/PD | 0 (0) | 6 (3) | 8 (6) | 2 (3) | 5 (4) | 3 (5) | ||
| Response at 2 y | ||||||||
| Observation arm | ||||||||
| CR/CRu | 15 (58) | 37 (43) | 44 (59) | .19 | 21 (43) | 43 (45) | 28 (59) | .17 |
| PR | 4 (15) | 11 (13) | 5 (6) | 5 (10) | 7 (7) | 7 (15) | ||
| SD/PD | 7 (27) | 38 (44) | 26 (35) | 20 (41) | 39 (41) | 11 (23) | ||
| Maintenance arm | ||||||||
| CR/CRu | 21 (72) | 72 (77) | 55 (80) | .61 | 43 (77) | 76 (73) | 27 (73) | .93 |
| PR | 4 (14) | 8 (8) | 3 (4) | 3 (5) | 9 (9) | 3 (8) | ||
| SD/PD | 4 (14) | 14 (15) | 11 (16) | 7 (13) | 14 (14) | 6 (16) | ||
| . | FCGR3A . | FCGR2A . | ||||||
|---|---|---|---|---|---|---|---|---|
| VV, n (%) . | VF, n (%) . | FF, n (%) . | P . | HH, n (%) . | HR, n (%) . | RR, n (%) . | P . | |
| Response to induction | ||||||||
| Whole series | ||||||||
| CR | 23 (34) | 75 (35) | 61 (35) | .86 | 41 (34) | 75 (34) | 38 (38) | .21 |
| CRu | 21 (31) | 69 (32) | 57 (33) | 31 (26) | 85 (38) | 28 (28) | ||
| CR/CRu | 44 (65) | 144 (67) | 118 (68) | 72 (60) | 160 (72) | 66 (66) | ||
| PR | 21 (31) | 61 (29) | 46 (26) | 43 (36) | 53 (24) | 30 (30) | ||
| SD/PD | 3 (4) | 8 (4) | 10 (6) | 5 (4) | 11 (4) | 5 (5) | ||
| R-CHOP regimen | ||||||||
| CR/CRu | 36 (67) | 121 (69) | 103 (71) | .29 | 62 (64) | 131 (72) | 60 (69) | .55 |
| PR | 18 (33) | 48 (27) | 34 (23) | 32 (33) | 43 (24) | 23 (26) | ||
| SD/PD | 0 (0) | 6 (3) | 8 (6) | 2 (3) | 5 (4) | 3 (5) | ||
| Response at 2 y | ||||||||
| Observation arm | ||||||||
| CR/CRu | 15 (58) | 37 (43) | 44 (59) | .19 | 21 (43) | 43 (45) | 28 (59) | .17 |
| PR | 4 (15) | 11 (13) | 5 (6) | 5 (10) | 7 (7) | 7 (15) | ||
| SD/PD | 7 (27) | 38 (44) | 26 (35) | 20 (41) | 39 (41) | 11 (23) | ||
| Maintenance arm | ||||||||
| CR/CRu | 21 (72) | 72 (77) | 55 (80) | .61 | 43 (77) | 76 (73) | 27 (73) | .93 |
| PR | 4 (14) | 8 (8) | 3 (4) | 3 (5) | 9 (9) | 3 (8) | ||
| SD/PD | 4 (14) | 14 (15) | 11 (16) | 7 (13) | 14 (14) | 6 (16) | ||
CR indicates complete response; CRu, unconfirmed complete response; PR, partial response; SD/PD, stable disease/progressive disease; and R-CHOP, rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone.
Response at the end of 2-year rituximab maintenance according to FCGR3A and FCGR2A alleles
Treatment maintenance provided an advantage for response in patients with all FCGR3A and FCGR2A genotypes (Table 2). As observed at the end of induction therapy, no difference in response rates was observed at 2 years from randomization in either the observation or the rituximab maintenance arms according to FCGR3A or FCGR2A genotypes. To better examine the effect of FCGR polymorphisms during rituximab maintenance and to determine whether the FCGR3A or FCGR2A genotype might influence response improvement, we focused our analysis on patients who attained PR after induction and achieved CR/CRu after rituximab maintenance. For FCGR3A, among the 57 patients in PR after induction therapy and then random assigned to the maintenance arm, 30 (57%) were in CR/CRu after 2 years of rituximab treatment, 15 (26%) remained in PR, 8 (14%) progressed, and 4 (7%) were not evaluated. The proportion of patients whose response status converted from PR to CR/CRu was not statistically different among the VV (n = 4; 40%), VF (n = 14; 52%), and FF (n = 12; 60%) alleles (P = .58). For FCGR2A, among the 56 patients in PR after induction, 30 (54%) obtained a CR/CRu after maintenance treatment, 15 (27%) remained in PR, 7 (13%) progressed, and 4 (7%) were not evaluated. The genotyping of patients whose response status converted from PR to CR/CRu showed no difference in allele distribution: HH (n = 11; 61%), HR (n = 14; 50%), or RR (n = 5; 50%; P = .84).
PFS and OS according to FCGR3A and FCGR2A alleles
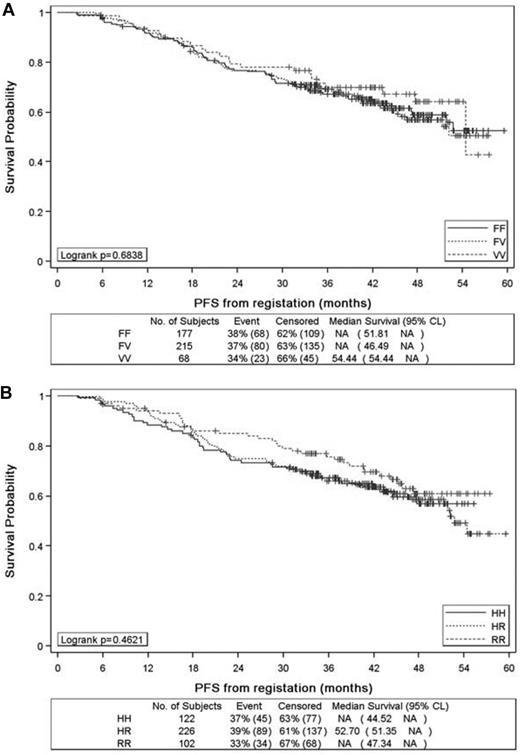
With a median follow-up of 44 months from the time of registration in the PRIMA study, the 3-year PFS rates for patients with FCGR3A VV, VF, and FF were 69.9% (95% CI, 57.2%-79.4%), 69.1% (95% CI, 62.3%-74.9%), and 67.3% (95% CI, 59.8%-73.7%), respectively (P = .68; Figure 1A). These rates were 67.1% (95% CI, 57.8%-74.8%), 65.9% (95% CI, 59.1%-71.7%), and 75.8% (95% CI, 66.0%-83.1%), respectively, for HH, HR, and RR carriers of FCGR2A SNP (P = .46; Figure 1B). No difference in median OS from registration was seen among the 3 allele groups in FCGR3A and FCGR2A SNPs (data not shown). Similar results were obtained when the analysis was restricted to patients treated with R-CHOP (data not shown).
Progression-free survival from the time of patient registration in the PRIMA study. Progression-free survival according to FCGR3A (A) and FCGR2A (B) alleles.
Progression-free survival from the time of patient registration in the PRIMA study. Progression-free survival according to FCGR3A (A) and FCGR2A (B) alleles.
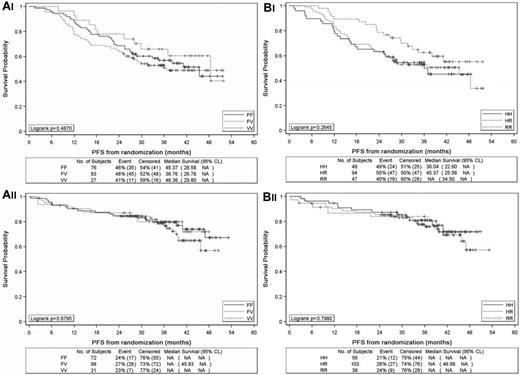
A comparison of PFS from random assignment with a median follow-up of 36 months was then performed among patients with FCGR3A VV, VF, and FF alleles. PFS rates for patients with VV, VF, and FF alleles in FCGR3A were 66.0% (95% CI, 44.8%-80.7%), 53.0% (95% CI, 42.2%-62.6%), and 56.8% (95% CI, 44.7%-67.3%), respectively, in the observation arm (P = .48; Figure 2Ai) and 79.8% (95% CI, 60.4%-90.4%), 78.6% (95% CI, 68.3%-85.9%) and 79.6% (95% CI, 67.9%-87.4%), respectively, in the rituximab maintenance arm (P = .46; Figure 2Aii). The 3-year PFS rate of VV carriers was thus slightly superior to that of non-VV carriers in the observation arm, but this difference was not significant (66% vs 54.6%; P = .31), and the curves were overlapping in the rituximab maintenance arm (79.8% vs 79%; P = .70).
Progression-free survival from the time of randomization between observation and rituximab maintenance in the PRIMA study. Progression-free survival according to FCGR3A alleles in the observation arm (Ai) and rituximab arm (Aii) and according to FCGR2A alleles in the observation arm (Bi) and rituximab arm (Bii).
Progression-free survival from the time of randomization between observation and rituximab maintenance in the PRIMA study. Progression-free survival according to FCGR3A alleles in the observation arm (Ai) and rituximab arm (Aii) and according to FCGR2A alleles in the observation arm (Bi) and rituximab arm (Bii).
The 3-year PFS rates according to FCGR2A HH, HR, and RR alleles were 54.4% (95% CI, 39.3%-67.2%), 52.7% (95% CI, 42.1%-62.3%), and 62.5% (95% CI, 46.7%-74.9%), respectively, in the observation arm (P = .26; Figure 2Bi) and 80.9% (95% CI, 67.3%-89.3%), 77.7% (95% CI, 67.7%-84.9%), and 83.9% (95% CI, 67.6%-92.4%), respectively, in the rituximab maintenance arm (P = .79; Figure 2Bii). These rates were not different between HH alleles and non-H alleles in the observation (54.4% vs 55.7%; P = .47) and the rituximab (80.9% vs 79.4%; P = .64) arms.
Influence of FCGR3A and FCGR2A alleles according to tumor bulk and FLIPI risk groups
Because previous reports showed that response to rituximab could be influenced by the presence of bulky disease,14 we analyzed whether FCGR SNPs had a differential effect for patients according to tumor bulk. We performed subanalyses in patients with or without bulky tumors (> or < 7 cm) as prospectively collected (this parameter was one of the inclusion criteria) and in the different prognostic groups of the FLIPI (≤ 1, 2, or 3-5 risk factors).2 The quality of response after the induction phase, PFS from registration and from random assignment for patients allocated to rituximab maintenance were not influenced by FCGR3A or FCGR2A genotypes in the different subgroups analyzed (data not shown).
Outcome according to FCGR3A and FCGR2A alleles stratified by lymphocyte counts
Because it was previously reported that a lymphocyte count at diagnosis ≥ 1.0 × 109/L influenced response and event-free survival in rituximab-treated patients,14 we hypothesized that FCGR polymorphism could play a distinct role in patients with low or high lymphocyte counts. We then analyzed the potential influence of FCGR3A and FCGR2A genotypes on response and outcome in those subgroups. At baseline, lymphocyte counts were available in 406 of the genotyped patients. In those patients, the median lymphocyte count was 1.26 × 109/L, with 259 patients presenting with a lymphocyte count ≥ 1.0 × 109/L and 147 patients with < 1.0 × 109/L. Responses to induction therapy or at the end of rituximab maintenance (in the 192 patients randomly assigned to this treatment arm) were not influenced by FCGR3A genotypes in the high and low lymphocyte count groups. Similar analyses were performed for FCGR2A SNP, and no difference in term of response was observed in the 2 lymphocyte count groups. The PFS from registration for FCGR3A was not different among the 3 genotypes in the low lymphocyte count group: the 3-year PFS rate was 70.6%, 68.6%, and 67.1% for VV, VF, and FF carriers, respectively (P = .47). No difference was observed in the high lymphocyte count group with a 3-year PFS rate of 73.9%, 69.8%, and 67.6% for VV, VF, and FF carriers, respectively (P = .78). Similar results were obtained according to the FCGR2A polymorphism. Evaluation of PFS from randomization in the rituximab arm also showed no difference among the different genotypes of FCGR3A and FCGR2A in patients with low or high lymphocyte count evaluated at the time of random assignment (data not shown).
Effect of FCGR genotypes on hematologic toxicities during treatment
A previous study has shown that FCGR3A polymorphisms could be associated with the degree of neutropenia manifest in the context of rituximab treatment after autologous transplantation.20 We therefore explored the relation between neutropenia and FCGR SNPs during induction treatment and rituximab maintenance. During induction treatment, 347 and 341 patients presented at least 1 episode of grade 2-4 neutropenia in the FCGR3A and FCGR2A genotyped cohorts, respectively. No difference in the frequencies of neutropenia was observed among FCGR3A VV (n = 48; 71%), VF (n = 168; 78%), and FF (n = 131; 74%) patients (P = .38) and among FCGR2A HH (n = 88; 72%), HR (n = 173; 77%), and RR (n = 80; 78%) patients (P = .27). When the analysis was restricted to grade 3/4 neutropenia during induction treatment, for FCGR3A SNP, 34 VV (50%) and 252 F carrier (FV or FF; 66%) patients presented at least 1 episode of grade 3 neutropenia (P = .02). However, this difference was not observed for the FCGR2A SNP. During rituximab maintenance, 7 (10%), 12 (6%), 10 (6%) patients with FCGR3A VV, VF, and FF genotypes, respectively, had at least 1 episode of grade 2-4 neutropenia (P = .34). Similarly, no difference was observed for FCGR2A: 11 HH (9%), 13 HR (6%), and 4 RR (4%) patients presented at least 1 grade 2-4 neutropenia (P = .13). Again, no difference was observed when the analyses were restricted to grade 3/4 neutropenia.
Discussion
In this large prospective substudy of the PRIMA trial, we found no influence of FCGR3A or FCGR2A polymorphisms on the response or outcome of patients with high tumor burden FL receiving a rituximab chemotherapy combination and rituximab maintenance. These observations were similar regardless of the tumor bulk or FLIPI category and were unaffected by the peripheral blood lymphocyte count. In contrast to the end points related to rituximab efficacy, we observed that FCGR3A polymorphisms were associated with the rate of grade 3-4 neutropenia during the combination of R-CT. Given the low number of patients developing this adverse event, we cannot exclude that this observation may be a chance association of limited clinical relevance. However, in accordance with previous reports, this result suggests that immune mechanisms mediated by natural killer (NK) cells may play a role in rituximab-induced neutropenia.20
The rationale to investigate these polymorphisms was based on previously published in vitro and in vivo studies. It is well established that the affinity of Fc region of hu-Ig G1 with FcγRIIIA is influenced by some SNPs in FCGR3A, for example, the presence of a V conferring a higher affinity than the presence of F at codon 158.21 In vitro, with therapeutic mAbs, these data were confirmed, with rituximab more efficiently binding and activating NK cells in VV carriers than in FF carriers.22,23 However, when analyzing the efficacy of B-cell lysis by NK cells obtained from healthy donors, it was found that differences in this lytic activity according to FCGR3A genotypes were only observed at low (< 0.01 μg/mL) rituximab concentrations.23 Serum concentrations of rituximab obtained in vivo, after infusions given every 3 weeks or every 2 months, usually substantially exceed these levels,24-27 although the concentration of antibody at tumor sites remained unknown. More recently, it was reported that some peripheral blood parameters reflecting NK-cell activation assessed 4 hours after an initial infusion of rituximab were lower in FF than in FV/VV patients.28 However, the significance of these findings with repeated rituximab infusions or in terms of clinical response remains unknown. Considering the other mechanisms of action of rituximab (apoptosis, complement-dependent cytotoxicity, phagocytosis) that could also eventually account for its therapeutic activity, a definite relation between NK-cell activation and the efficacy of the antibody in vivo is still lacking.
On the basis of this biologic rationale, many retrospective studies in patients treated with therapeutic IgG1 mAbs analyzed the correlation between FCGR polymorphisms and treatment response or outcome.12,13,29-31 Considering specifically FL, 8 studies have previously examined these correlations.12,13,15,32-36 The first demonstration of the variability in the quality of response to rituximab treatment according to FCGR3A polymorphism was derived from analysis of a cohort of 49 patients with low tumor burden FL receiving 4 infusions of rituximab as first-line treatment.12 Significantly higher clinical and molecular response rates were found in VV carriers, and a possible improvement in PFS was suggested. In contrast, no influence of the FCGR2A polymorphism was found in this study.12 A second cohort of 87 patients receiving single-agent rituximab was retrospectively analyzed for the effect of FCGR3A and FCGR2A polymorphisms and reported their influence on both response rates and PFS.13 In the context of a prolonged rituximab treatment schedule (4 weekly infusions followed by 4 infusions administered every 2 months), again used as single agent, response and event-free survival were improved mainly for patients with FCGR3A VV genotypes.14,34 This study of 171 patients also established that FCGR3A status was an independent prognostic factor for event-free survival.14 These latter studies were retrospective and included both untreated and previously treated patients. Preliminary results obtained in the prospective study that assessed rituximab single agent followed by 2 years of rituximab maintenance in patients with low tumor burden FL were recently reported.33 In this study, FCGR3A, FCGR2A, and FCGR2B SNPs were not found to influence the outcome of patients receiving rituximab maintenance.33 When considering rituximab plus chemotherapy combinations, 3 prior retrospective reports (including 75-102 patients) failed to find any effect of FCGR2A and FCGR3A status on prognosis.15,32,36 However, discordant results were reported from a prospective clinical trial that included 76 patients treated with chemotherapy in combination with either rituximab or I-131 tositumomab, whereby FCGR3A alleles were associated with OS.34 Two additional studies should be mentioned in this context. First, the outcome of patients with FL treated with chemotherapy alone or under watchful waiting does not appear to be affected by FCGR3A or FCGR2A polymorphisms,34,37 clearly indicating that the effects of FcγR variability on outcome are restricted to patients receiving mAb therapy. Second, the identification of FCGR2A polymorphism effect may potentially only reflect the linkage disequilibrium between the FCGR3A rs396991 and the FCGR2A rs1801274 SNPs rather than a true biologic mechanism related to the affinity of the FcγRIIA allotypes.38,39
The present results represent the largest cohort prospectively analyzed and clarify this debate. The characteristics and outcome of the 460 patients who provided consent for genotyping did not meaningfully differ from patients of the whole PRIMA study. The relative proportion of FCGR3A and FCGR2A SNP genotypes, mainly from Europe and Australia, was in accordance with other studies and with the Hardy-Weinberg equilibrium. Because the PRIMA study design included 2 steps, we were able to analyze the effects of FCGR SNPs both after R-CT induction and also for those patients with responsive disease who received rituximab maintenance. In accordance with other reports from smaller retrospective studies,15,32,36 our data indicate that FCGR3A and FCGR2A polymorphisms have no effect on response rate and PFS for patients treated with rituximab chemotherapy combinations. Therefore, the effects of FcγR polymorphisms on the therapeutic activity of rituximab are only observed when this antibody is used as a single agent. One possible explanation is that chemotherapy agents may alter immune mechanisms such as antibody-dependent cellular cytotoxicity (ADCC) in those patients. However, this does not preclude the therapeutic activity of the mAbs when associated with chemotherapy.40 These results are also in line with those observed in the context of other B-cell malignancies, such as diffuse large B-cell lymphoma or chronic lymphocytic leukemia.41,42
More surprising in this regard was the lack of correlation between FCGR genotypes and response after maintenance or PFS in those patients who received 2 years of rituximab maintenance. Again, the effects of previous chemotherapy may partly account for these observations, by altering the ADCC effector functions. Our analysis of lymphocyte count used levels at registration and at time of randomization and is limited by the lack of evaluation of lymphocyte subsets, such as NK lymphocytes, for instance.27 Moreover, lymphocyte subsets are not the sole immune effectors of anti-CD20 mAbs; the role of macrophages also being important.43,44 Another hypothesis is that the schedule of rituximab administration (1 infusion of 375 mg/m2 every 2 months for 24 months) abolishes the variability of the therapeutic effects related to FCGR polymorphisms. With this schedule, sustained levels of rituximab are probably achieved between each infusion, probably leading to a saturation of FCγR binding.24,45 It is also conceivable that the efficacy of rituximab according to FCGR polymorphisms may be observed in patients with low tumor burden, not represented in the present study; however, we did not observe any influence of FCGR genotypes in patients without tumor bulk.
In conclusion, this large series of patients with FL treated in a prospective trial indicates that the outcome of R-CT followed by 2-year rituximab maintenance is not influenced by FCGR3A and FCGR2A genotypes. Other genetic determinants of anti-CD20 mAb activity may also need to be examined in the future. New anti-CD20 mAbs are currently under development to improve ADCC, complement-dependent cytotoxicity, or induction of programmed cell death.10 In an in vitro study on peripheral blood mononuclear cells of healthy donors, low fucose anti-CD20 IgG1 enhanced ADCC compared with rituximab and was independent of FCGR3A SNP.46 Clinically, a phase 1/2 study in relapsed or refractory B-cell malignancies has shown that objective responses to a new humanized IgG1 with afucosylated Fc appears to be independent of FCGR3A SNP.47 Our results suggest that ADCC optimization may be more relevant to explore when using anti-CD20 antibodies alone or in combination with other immunomodulating agents48 that may enhance cellular cytotoxicity.
The online version of this article contains a data supplement.
Presented in part at the 11th International Conference on Malignant Lymphoma (ICML), Lugano, Switzerland, June 17, 2011.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank M. D. Reynaud for editorial assistance; Celine Mabon for technical assistance; the GELARC (Groupe d'Etude des Lymphomes de l'Adultes Recherche Clinique) team, and especially Anne-Laure Borrel and Delphine Germain for study management as well as Bénédicte Gelas-Dore for statistical analyses.
This work was supported in part by a grant from the Institut National du Cancer (INCa, Paris, France) French Ministry of Health.
Authorship
Contribution: H.G. designed the research, performed the research, analyzed the data, and wrote the manuscript; G.S. designed the research, analyzed the data, and wrote the manuscript; H.G., G.C., J.F.S., M.-H.D.-L., F.O., P.S., A.P., P.B., R.B., A.S., J.D., O.C., J.V.C., A.D., F.J., A.V., P.D., and G.S. provided study materials or patients, collected and assembled data, and approved the final manuscript.
Conflict-of-interest disclosure: G.C., G.S., and J.F.S. have received compensation for consultancy, advisory board, and talks from Roche. O.C. has received compensation for consultancy. The remaining authors declare no competing financial interests.
Correspondence: Gilles Salles, Service d'Hématologie, Centre Hospitalier Lyon-Sud, 165, chemin du Grand Revoyet, 69495 Pierre-Bénite Cedex, France; e-mail: gilles.salles@lyon-chu.fr.