Abstract
The chemokine receptor CXCR4 is a critical regulator of cell migration and serves as a coreceptor for HIV-1. The chemokine stromal cell derived factor-1, also known as CXCL12, binds to CXCR4 and exerts its biologic functions partly through the small guanosine triphosphate hydrolase (GTPase) Rac1 (ras-related C3 botulinum toxin substrate 1). We show in different cell types, including CD34+ hematopoietic stem and progenitor cells, that inhibition of Rac1 causes a reversible conformational change in CXCR4, but not in the related receptors CXCR7 or CCR5. Biochemical experiments showed that Rac1 associates with CXCR4. The conformational change of CXCR4 on Rac1 inhibition blocked receptor internalization and impaired CXCL12-induced Gαi protein activation. Importantly, we found that the conformation adopted by CXCR4 after Rac1 inhibition prevents HIV-1 infection of both the U87-CD4-CXCR4 cell line and of primary peripheral blood mononuclear cells. In conclusion, our data show that Rac1 activity is required to maintain CXCR4 in the responsive conformation that allows receptor signaling and facilitates HIV-1 infection; this implies that Rac1 positively regulates CXCR4 function and identifies the Rac1-CXCR4 axis as a new target for preventing HIV-1 infection.
Introduction
The 7 transmembrane-spanning Gαi protein-coupled chemokine receptor CXCR4 and its ligand CXCL12 are involved in the regulation of vital processes involving cellular migration, such as angiogenesis, neuronal development, and hematopoietic stem cell (HSC) and immune cell trafficking. CXCL12 or CXCR4 deficiency in mice leads to embryonic lethality, underscoring the importance of this chemokine/receptor pair.1,2 CXCR4 is also implicated in pathologic conditions, including tumor cell proliferation and metastasis,3 and notably, serves as a coreceptor for T-cell tropic HIV-1 strains.4
Ligand binding to CXCR4 induces a conformational change of the receptor, which then can act as a guanine nucleotide exchange factor for the heterotrimeric Gαi protein, facilitating the exchange of guanosine diphosphate (GDP) for guanosine triphosphate (GTP) on the Gαi subunit.5 These events initiate the activation of multiple signaling pathways leading to the generation of a chemotactic response toward a CXCL12 gradient in various types of cells.6
Cellular motility, mediated by CXCR4, relies on the activation and signaling by the Rho family of GTPases, which control cytoskeletal dynamics, providing both protrusive force at the front and contraction at the rear of the cell. The best known members of the Rho family are RhoA (Ras homologue gene family member A), that stimulates myosin-based contractility, and CDC42 (cell division cycle 42) and Rac1 (ras-related C3 botulinum toxin substrate 1), that induce actin polymerization and membrane protrusions at the leading edge.7 Rho GTPases are thus at the center of actin regulation in migrating cells and act as molecular switches, cycling between an active GTP-bound state and an inactive GDP-bound state. This cycle is regulated by guanine nucleotide exchange factors and GTPase activating proteins.8
The initiation of directional migration in response to CXCL12 is dependent on the level of cell surface expression of the chemokine receptor CXCR4, as well as the cell's ability to properly initiate signaling on ligand binding. Similarly, T-cell tropic HIV-1 infection depends on the surface expression of CXCR4. Initial binding of the viral envelope glycoprotein 120 (gp120) is mediated by CD4. This step induces a structural rearrangement of gp120, which exposes a binding site for CXCR4.9 Subsequently, gp120 binding to CXCR4 induces viral fusion and entry into host cells. In vitro studies have shown that gp120 binding triggers CXCR4 signaling and presumably creates an intracellular environment that facilitates postentry HIV-1 infection events.10,11
It is known that cells display a heterogeneous set of CXCR4 conformations12 that have been associated with different Gαi activation patterns. Moreover, certain conformations generated inactive receptors.13 Thus, the conformation adopted by CXCR4 determines ligand binding and/or signaling capacity and possibly affects coreceptor function.14
The molecular mechanisms that regulate CXCR4 conformation at steady state conditions are unknown. Here, we demonstrate that Rac1 is specifically involved in regulating the conformation of CXCR4 in several human cell types, including CD34+ hematopoietic stem and progenitor cells (HSPCs), thereby controlling signaling efficiency of the receptor. Furthermore, the conformation adopted by CXCR4 after Rac1 inhibition blocked HIV-1 infection, most probably by interfering with virus binding and subsequent entry into host cells. Our findings are the first to describe the involvement of a GTPase in the regulation of the CXCR4 responsive conformation, which represents a previously unrecognized role for Rac1 in Gαi protein-coupled receptor homeostasis.
Methods
Dominant negative peptides for and pharmacologic inhibition of Rac1
Cell permeable peptides containing the amino acid sequence of the C–terminal region of Rac1 (CPPPVKKRKRK) and CDC42 (LEPPEPKKSRR) fused to a protein transduction domain (PTD) or a control peptide that only encodes the PTD (control) were produced by F-moc protein synthesis (Department of Peptide Synthesis, Netherlands Cancer Institute, Amsterdam, The Netherlands). Rac1 C–terminal mutant peptides were produced by replacing the 3 prolines, that is aa 179-181 (CAAAVKKRKRK), or the RKR sequence, that is aa 185-187 (CPPPVKKAAAK), by alanine residues (see Figure 1).
Cells were harvested in Iscove modified Dulbecco medium (IMDM; Lonza) containing 0.25% wt/vol BSA (Sigma-Aldrich) named assay medium and treated with 100 μg/mL or 200 μg/mL peptide for 30 minutes. Pharmacologic inhibition of Rac1 was performed with NSC23766 (Calbiochem) or EHT1864 (Sigma-Aldrich) for 1 hour at concentrations of 25μM or 50μM in assay medium. Subsequently, cells were washed with PBS (Fresenius Kabi) containing 0.5% wt/vol BSA named FACS medium. The DNA-binding dye, ToPro3 (Invitrogen), was used to determine cytotoxicity. CXCL12 (PeproTech) was used at a final concentration of 100 ng/mL for 30 minutes as a positive control for responsiveness of the CXCR4 receptor on the cell surface.
Peptide washout experiment
Cells were treated with the Rac1 C–terminal peptide as previously described and subsequently washed twice with assay medium to remove the peptide. Thereafter, the cells were placed at 37°C. At different time points (0, 15, 30, and 60 minutes) cells were transferred to a 96-well V-bottom plate, washed with FACS medium and fixed with 2% formaldehyde for 30 minutes at room temperature. Subsequently, cells were washed with FACS medium and cell surface expression levels of CXCR4 were determined using specific CXCR4 monoclonal antibodies (mAbs; see supplemental Methods for specifications, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) and flow cytometry.
All flow cytometry measurements were performed using the FACS CANTO, FACS LSR II, or FACS LSR II HTS (BD Biosciences), and results were analyzed with BD FACSDiva Version 6.1 software (BD Biosciences).
Antibody feeding experiment
In antibody feeding experiments, HL60 cells were stained with specific CXCR4 mAbs at 4°C and subsequently washed with FACS medium to remove unbound antibodies. Thereafter, the cells were incubated with assay medium, the control peptide or the Rac1 peptide for periods ranging from 5 to 45 minutes at 4°C or 37°C, or with NSC23766 for 1 hour at 37°C. Subsequently, the cells were washed with FACS medium and the signal of the anti-CXCR4 antibody was determined by flow cytometry.
Peptide pull-down assay
HeLa cells were transiently transfected with the HA-CXCR4 construct and maintained for 24 hours. The cells were washed twice with ice-cold PBS (supplemented with 1mM CaCl2 and 0.5mM MgCl2) and lysed in NP-40 lysis buffer (ie, 50mM Tris-HCl pH 7.5, 100mM NaCl, 10mM MgCl2, 10% glycerol, and 1% NP-40) supplemented with protease inhibitors (Complete mini EDTA; Roche), centrifuged at 20 000g for 10 minutes at 4°C. The supernatant was precleared by incubation with streptavidin-coated beads (Sigma-Aldrich) for 1 hour at 4°C. The precleared lysate was then incubated with the indicated Rho GTPase C–terminal peptides, a peptide representing the effector domain (ED) of Rac1 (amino acids 17-32)15 or a control peptide (8 μg) in the presence of streptavidin-coated beads (blocked with 4% BSA) at 4°C for 1 hour with rotation. Initial lysates and pull-down samples were analyzed by SDS-PAGE and immunoblotting using mouse anti-HA (Sigma-Aldrich) and HRP-coupled secondary antibodies.
Immunoprecipitation
HeLa cells were transiently transfected with HA-CXCR4 and myc-Rac1Q61L or myc-Rac1T17N constructs and maintained for 24 hours. After lysing the cells as described in “Peptide pull-down assay,” the supernatant of the lysate was precleared by incubation with protein G-agarose beads for 1 hour at 4°C. The precleared lysates were then incubated with monoclonal anti-HA antibodies and protein G-agarose beads (blocked with 4% BSA) at 4°C for 5 hours. Samples were then washed 5 times with lysis buffer. Immunoprecipitated proteins were eluted by adding sodium dodecyl sulfate (SDS) sample buffer and heating at 50°C for 20 minutes. Protein association was assayed by SDS-PAGE and immunoblotting using rabbit anti-HA and rabbit anti-myc antibodies (both from Sigma-Aldrich) and HRP-coupled secondary antibodies.
CRE-luciferase reporter gene assay
HEK293T cells were transiently transfected using polyethyleninine (PEI) with plasmids encoding for CXCR4 and the cyclic AMP responsive element (CRE) coupled to the luciferase gene (CRE-Luc).16 Briefly, 1 μg of CXCR4 and 1 μg CRE-Luc plasmids were transfected per 106 HEK293T cells and seeded onto a white 96-well plate (Greiner). Two days posttransfection, the culture medium was discarded and cells were stimulated with DMEM supplemented with 0.5% wt/vol BSA, forskolin (3μM), and a concentration range of CXCL12 varying from 1pM to 100nM in the absence or presence of the Rac1 inhibitors. After 8 hours of incubation at 37°C, the stimulation medium was discarded and the cells were lysed in the presence of D-luciferin (Duchefa Biochemi BV). Luminescence was measured on a Wallac Victor plate reader2 (Perkin Elmer).
Human immunodeficiency viruses and infection assays
Single round luciferase reporter virus was produced by cotransfection of the pNL4-3.Luc.R-E-construct17 and constructs expressing either BaL-26 envelop or HxB2 envelop in HEK293T cells using calcium phosphate. Infectious virus was harvested at 48 and 72 hours after transfection and filtered through a 0.22-μm filter. Virus titers were quantified by determination of the 50% tissue culture infectious dose (TCID50) on U87 cells expressing CD4 and CCR5 (U87-CD4-CCR5) or CXCR4 (U87-CD4-CXCR4). After 48 hours, luciferase substrate (0.83mM ATP, 0.83mM d-luciferin, 18.7mM MgCl2, 0.78μM Na2H2P2O7, 38.9mM Tris-HCl pH 7.8, 0.39% vol/vol glycerol, 0.03% vol/vol Triton X-100, and 2.6μM dithiothreitol) was added. Luminesence was measured using the Centro LB 960 (Berthold Technologies).
HIV-1 variant NL4-3 was produced by transfection of the full-length construct in HEK293T cells using calcium phosphate. Infectious virus was harvested at 48 and 72 hours after transfection and filtered through a 0.22-μm filter. Virus titers were quantified by determination of the 50% tissue culture infectious dose on phytohemagglutinin (PHA)–stimulated peripheral blood mononuclear cells (PBMCs) using an in-house p24 ELISA.18
For the single round luciferase reporter HIV-1 infection assay, U87 cells expressing CD4, and CCR5 or CXCR4 were plated at a cell density of 6000 cells per well and cultured for 24 hours. Next, the cells were inoculated with 200 TCID50 BaL-26 or HxB2 env pseudotyped single round luciferase virus and 48 hours later, the infectivity of the virus was analyzed by the luciferase assay. During and after inoculation, the cells were treated with different concentrations of NSC23766 as indicated.
For the HIV-1 products qPCR, PHA-stimulated PBMCs (0.5 × 106) were inoculated with 250 TCID50 DNase treated NL4-3 in the presence or absence of 50μM NSC23766. After 48 hours, cells were harvested for DNA isolation to be used in the qPCR studies.
Calculations and statistical analysis
The percentage of CXCR4 surface expression was calculated by normalizing the fluorescence intensity measurements on the mean of the untreated condition measurements. Measurements of both treated and untreated conditions were divided by the mean of the untreated conditions (×100%), giving rise to a mean of 100% ± SEM for the control. The isotype control background was not subtracted from the specific fluorescence intensity signal.
pEC50 was determined by fitting a sigmoidal dose-response curve using GraphPad Prism 5 software.
Significance of differences was determined in all experiments with a 2-sided unpaired Student t test. Two-sided P values smaller than .05 were considered significant.
For further information on the materials and methods used, see supplemental Methods.
Results
Inhibition of Rac1 specifically decreases CXCR4 signal on the cell surface
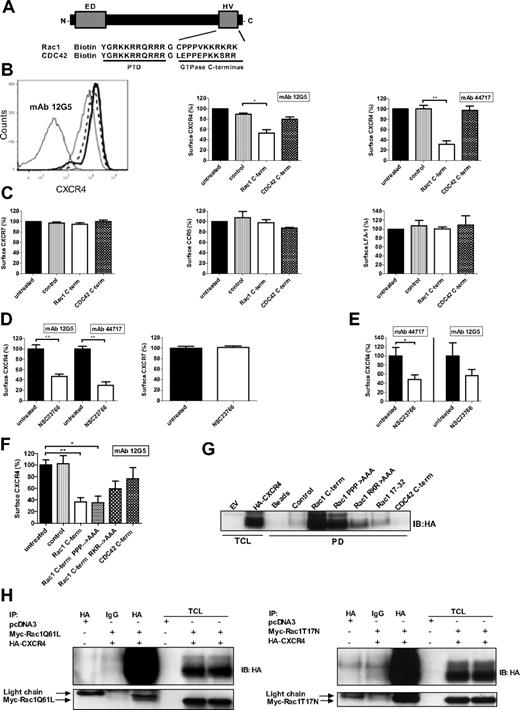
To investigate whether Rac1 modulates CXCR4 function, we determined the effect of Rac1 inhibition on CXCR4 expression. We opted for a peptide-mediated approach and the use of pharmacologic inhibitors of Rac1. Both methods ensure a rapid inhibition of Rac1, in contrast to the expression of a dominant-negative (DN) construct of Rac1. Inhibition of Rac1 with a biotinylated Rac1 C–terminal peptide (Figure 1A),19 significantly reduced the CXCR4 cell surface signal detected by mAb clones 12G5 (Figure 1B left and middle panels) and 44717 in the myeloid HL60 cell line that expresses CXCR4 endogenously (Figure 1B right panel). The results obtained with these CXCR4 mAb clones were consistent and the inhibitory peptide did not affect cell viability (data not shown). Importantly, peptide-mediated inhibition of a related GTPase, CDC42, did not cause a significant decrease in the CXCR4 surface signal, indicating the specificity of Rac1 for the regulation of steady-state CXCR4 expression (Figure 1B-C). The cell surface expression of the alternative CXCL12 receptor CXCR7, the related chemokine receptor CCR5 and the unrelated adhesion molecule LFA-1 were not reduced when HL60 cells were treated with the Rac1 inhibitory peptide (Figure 1C). Thus, Rac1 specifically controls CXCR4 exposure.
Rac1 inhibition causes a decrease of CXCR4 surface signal and Rac1 associates with CXCR4. (A) Schematic representation of the Rho-like GTPase C–terminal peptides fused to a protein transduction domain as used in this study. (B) HL60 cells were incubated with the Rac1 or CDC42 inhibitory peptides and CXCR4 expression was detected by flow cytometry using the fluorescently labeled mAb 12G5 or 44717. The panels show a histogram overlay (left) and bar graphs (middle and right) representing the CXCR4 surface signal. Histogram: thin solid line: isotype control mAb, long dashed line: untreated, thick solid line: control peptide, dashed line: CDC42 C–terminal peptide, and dotted line: Rac1 C–terminal peptide. The CXCR4 surface signal in the untreated condition and control peptide treated condition completely overlap (n = 3). (C) HL60 cells were incubated with the Rac1 or CDC42 inhibitory peptides and CXCR7, CCR5, and LFA-1 surface signal were measured by flow cytometry (n = 3). (D) U937 cells were treated with 50μM NSC23766 and the CXCR4 (detected by mAbs 12G5 and 44717), and CXCR7 surface signals were measured by flow cytometry (n = 3). (E) CD34+ cells isolated from CB were treated with NSC23766 and the CXCR4 signal (detected by mAb 12G5 or 44717) was measured by flow cytometry (n = 5). (F) HEK293T cells exogenously expressing CXCR4-GFP were treated with the Rac1 (mutant) C–terminal peptides or the CDC42 C–terminal peptide and the CXCR4 surface signal (detected by mAb 12G5) was measured on the GFP-positive cells by flow cytometry (n = 3). (G) Pull-down (PD) experiment was performed using lysates from HeLa cells exogenously expressing HA-CXCR4 with beads only, a control peptide, wild-type and mutant Rac1 C–terminal peptides, the Rac1 17-32 ED peptide and the CDC42 C–terminal peptide. Association of CXCR4 was detected by immunoblotting (IB) with an HA-specific monoclonal antibody (representative example of 2 independent experiments). (H) Immunoprecipitation assays were performed in HeLa cells transfected with an empty vector (pcDNA3) or with HA-CXCR4 and cotransfected with myc-Rac1Q61L or myc-Rac1T17N. Immunoprecipitation of the receptor was performed using an HA-specific antibody or an IgG control antibody and immunoblotting was performed with an HA-specific or a myc-specific antibody (representative example of 2 independent experiments). ED indicates effector domain of Rac1; HV, hypervariable domain of Rac1; PTD, protein transduction domain; Rac1 PPP→AAA, Rac1 RKR→AAA, Rac1 C–terminal peptide mutants where the 3 prolines, or RKR sequence were replaced by alanine residues, respectively; Rac 17-32, effector domain; TCL, total cell lysates; and EV, empty vector. Bars show the median (B-C) and the mean (D-F) fluorescence intensity determined by flow cytometry and expressed as percentage ± SEM compared with untreated or to control conditions (*P < .05, **P < .01).
Rac1 inhibition causes a decrease of CXCR4 surface signal and Rac1 associates with CXCR4. (A) Schematic representation of the Rho-like GTPase C–terminal peptides fused to a protein transduction domain as used in this study. (B) HL60 cells were incubated with the Rac1 or CDC42 inhibitory peptides and CXCR4 expression was detected by flow cytometry using the fluorescently labeled mAb 12G5 or 44717. The panels show a histogram overlay (left) and bar graphs (middle and right) representing the CXCR4 surface signal. Histogram: thin solid line: isotype control mAb, long dashed line: untreated, thick solid line: control peptide, dashed line: CDC42 C–terminal peptide, and dotted line: Rac1 C–terminal peptide. The CXCR4 surface signal in the untreated condition and control peptide treated condition completely overlap (n = 3). (C) HL60 cells were incubated with the Rac1 or CDC42 inhibitory peptides and CXCR7, CCR5, and LFA-1 surface signal were measured by flow cytometry (n = 3). (D) U937 cells were treated with 50μM NSC23766 and the CXCR4 (detected by mAbs 12G5 and 44717), and CXCR7 surface signals were measured by flow cytometry (n = 3). (E) CD34+ cells isolated from CB were treated with NSC23766 and the CXCR4 signal (detected by mAb 12G5 or 44717) was measured by flow cytometry (n = 5). (F) HEK293T cells exogenously expressing CXCR4-GFP were treated with the Rac1 (mutant) C–terminal peptides or the CDC42 C–terminal peptide and the CXCR4 surface signal (detected by mAb 12G5) was measured on the GFP-positive cells by flow cytometry (n = 3). (G) Pull-down (PD) experiment was performed using lysates from HeLa cells exogenously expressing HA-CXCR4 with beads only, a control peptide, wild-type and mutant Rac1 C–terminal peptides, the Rac1 17-32 ED peptide and the CDC42 C–terminal peptide. Association of CXCR4 was detected by immunoblotting (IB) with an HA-specific monoclonal antibody (representative example of 2 independent experiments). (H) Immunoprecipitation assays were performed in HeLa cells transfected with an empty vector (pcDNA3) or with HA-CXCR4 and cotransfected with myc-Rac1Q61L or myc-Rac1T17N. Immunoprecipitation of the receptor was performed using an HA-specific antibody or an IgG control antibody and immunoblotting was performed with an HA-specific or a myc-specific antibody (representative example of 2 independent experiments). ED indicates effector domain of Rac1; HV, hypervariable domain of Rac1; PTD, protein transduction domain; Rac1 PPP→AAA, Rac1 RKR→AAA, Rac1 C–terminal peptide mutants where the 3 prolines, or RKR sequence were replaced by alanine residues, respectively; Rac 17-32, effector domain; TCL, total cell lysates; and EV, empty vector. Bars show the median (B-C) and the mean (D-F) fluorescence intensity determined by flow cytometry and expressed as percentage ± SEM compared with untreated or to control conditions (*P < .05, **P < .01).
To confirm these results, we used the pharmacologic Rac1-guanine nucleotide exchange factor inhibitor, NSC23766, which does not affect the function of CDC42 or RhoA.20 This inhibitor did not induce cellular toxicity in our experimental conditions (data not shown). In accordance with the previous findings, NSC23766-mediated Rac1 inhibition significantly reduced CXCR4 cell surface signal on U937 cells (Figure 1D left panel) and HL60 cells (data not shown). CXCR7 expression on these cells was not affected, again showing the specificity of Rac1 for CXCR4 regulation (Figure 1D right panel). The effect of Rac1 inhibition on the CXCR4 surface signal was further confirmed on human cord blood (CB)–derived CD34+ cells (Figure 1E). The same results were also observed with the pharmacologic Rac1 inhibitor EHT1864 (data not shown). These results indicate that Rac1 specifically regulates the CXCR4 signal on resting cells.
The Rac1 C-terminus contains a proline-rich domain and a polybasic domain, which are both involved in the binding of specific effector proteins.19 To examine whether these domains are involved in regulating CXCR4 expression, we used the same Rac1 peptide, but now with substitutional mutations of the proline-rich amino acid sequence or the polybasic amino acid sequence in HEK293T cells transiently expressing CXCR4. The Rac1 peptide with a mutated proline-rich sequence decreased the CXCR4 signal to the same extent as the wild-type Rac1 peptide. The Rac1 peptide with a mutated polybasic region resulted in a less prominent decrease of the CXCR4 signal suggesting the involvement of the polybasic binding motif. As observed in other cell types, peptide-mediated inhibition of CDC42 in HEK293T cells exogenously expressing CXCR4 did not cause a significant decrease in CXCR4 surface expression (Figure 1F). Overall, these data show that Rac1 specifically regulates CXCR4 expression in distinct cell types and that the polybasic region of the Rac1 C-terminus is at least partially required for Rac1 to exert this function.
To asses whether Rac1 and CXCR4 associate, we performed a streptavidin-based pull-down assay using the different biotinylated peptides previously described or the Rac1 peptide representing part of the ED (aa 17-32)15 in lysates of HeLa cells expressing hemagglutinin (HA)–tagged CXCR4. We found that CXCR4 clearly interacts with the Rac1 C–terminal peptide independently of the proline-rich domain. Binding was partially abolished when the polybasic region was mutated and we did not observe an interaction between HA-CXCR4 and the CDC42 C–terminal peptide or the Rac1 ED peptide (Figure 1G). These results correlate with the reduced effect of the polybasic mutant Rac1 C–terminal peptide on the CXCR4 surface signal compared with the wild-type or the proline mutant Rac1 C–terminal peptide as observed in Figure 1F. Next, to investigate the interaction between CXCR4 and full-length Rac1, we performed immunoprecipitation experiments in HeLa cells transfected with HA-CXCR4 and the myc-tagged active mutant of Rac1 (Q61L) or the myc-tagged DN mutant of Rac1 (T17N). Our results show that both myc-Rac1Q61L and myc-Rac1T17N coimmunoprecipitated with CXCR4 (Figure 1H). Together, these data show that Rac1, via its C-terminus, associates with CXCR4 independently of the GDP-GTP loading state of the GTPase and of agonist stimulation of the receptor.
Rac1 inhibition causes a conformational change of CXCR4
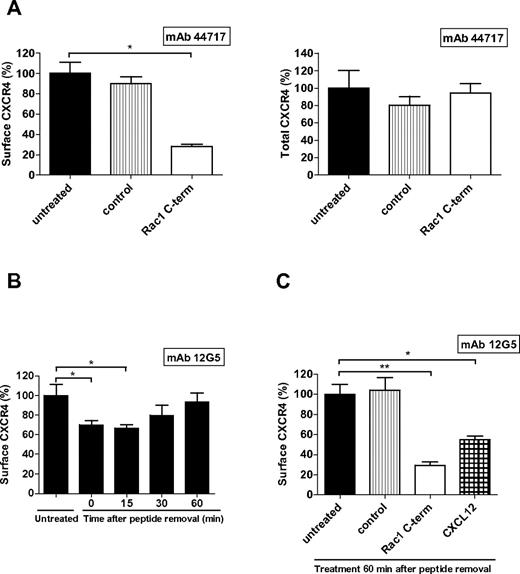
To investigate the fate of CXCR4 after inhibition of Rac1, HL60 cells were treated with the inhibitory Rac1 peptide after which surface and total CXCR4 expression were measured. Inhibition of Rac1 caused a significant decrease of CXCR4 detected on the cell surface (Figure 2A left panel), whereas the total levels remained unaffected compared with control cells (Figure 2A right panel). To determine whether the effect of Rac1 inhibition on CXCR4 expression was reversible, HL60 cells were first treated with the Rac1 inhibitory peptide, which was subsequently removed by washing the cells. Thereafter, surface levels of CXCR4 were measured after various time intervals during 60 minutes. The CXCR4 signal on the cell surface was still significantly decreased at 15 minutes after peptide removal, but returned to baseline levels compared with untreated cells after 60 minutes (Figure 2B). Moreover, at this time point CXCR4 was responsive, as shown by CXCL12-induced receptor internalization measured by a decrease in receptor surface expression. In addition, treating the cells again with the Rac1 inhibitory peptide resulted once more in a decrease of CXCR4 surface signal (Figure 2C). Together these results show that inhibition of Rac1 causes a reversible decrease of the CXCR4 surface signal, which is not because of CXCR4 degradation.
The effect of Rac1 inhibition on CXCR4 is reversible. (A) HL60 cells were incubated with the Rac1 inhibitory peptide and then surface CXCR4 (left) or total CXCR4 (right) were measured using mAb 44717 by flow cytometry (n = 3). (B) HL60 cells were incubated with the Rac1 inhibitory peptide, washed and placed at 37°C and CXCR4 surface signal (detected by mAb 12G5) was determined after different time periods by flow cytometry (n = 4). (C) One hour after washout of the Rac1 inhibitory peptide, HL60 cells were incubated again with the inhibitory peptide or with CXCL12 and the CXCR4 surface signal (as detected by mAb 12G5) was determined by flow cytometry (n = 3). Bars show the mean (A) and median (B-C) fluorescence intensity determined by flow cytometry and expressed as percentage ± SEM compared with untreated conditions (*P < .05, **P < .01).
The effect of Rac1 inhibition on CXCR4 is reversible. (A) HL60 cells were incubated with the Rac1 inhibitory peptide and then surface CXCR4 (left) or total CXCR4 (right) were measured using mAb 44717 by flow cytometry (n = 3). (B) HL60 cells were incubated with the Rac1 inhibitory peptide, washed and placed at 37°C and CXCR4 surface signal (detected by mAb 12G5) was determined after different time periods by flow cytometry (n = 4). (C) One hour after washout of the Rac1 inhibitory peptide, HL60 cells were incubated again with the inhibitory peptide or with CXCL12 and the CXCR4 surface signal (as detected by mAb 12G5) was determined by flow cytometry (n = 3). Bars show the mean (A) and median (B-C) fluorescence intensity determined by flow cytometry and expressed as percentage ± SEM compared with untreated conditions (*P < .05, **P < .01).
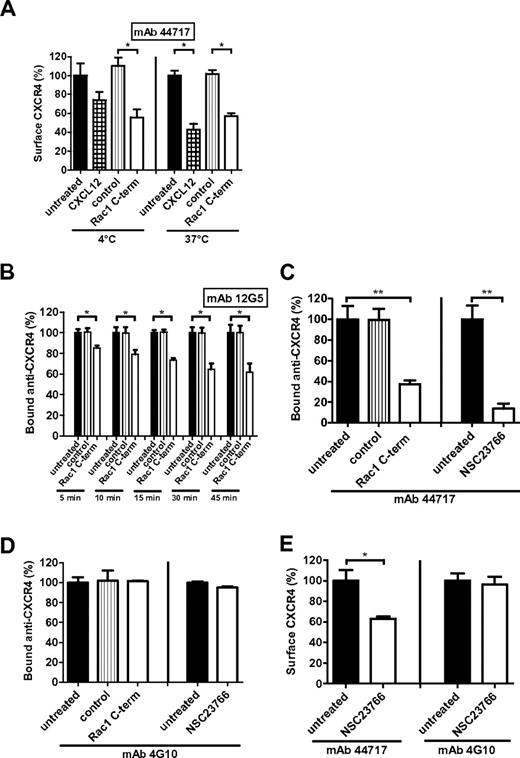
To determine whether Rac1 inhibition enhances CXCR4 internalization, HL60 cells were incubated with the Rac1 inhibitory peptide or NSC23766 at 4°C, at which temperature vesicular trafficking, including internalization of membrane proteins, is abrogated. As a control, cells were treated with CXCL12. CXCL12-induced internalization of CXCR4 was severely impaired at 4°C compared with 37°C, indicating that receptor internalization is indeed abolished at low temperatures (Figure 3A). Peptide-mediated inhibition of Rac1 at 4°C, however, resulted in the same CXCR4 surface signal decrease as observed at 37°C (Figure 3A). Incubation of the cells with NSC23766 at 4°C also resulted in a significant decrease of the CXCR4 surface signal (data not shown). Therefore, it is unlikely that the decrease of the CXCR4 surface signal on Rac1 inhibition is because of enhanced endocytosis.
Rac1 inhibition alters CXCR4 conformation. (A) HL60 cells were incubated with CXCL12 or the Rac1 inhibitory peptide at 4°C or 37°C and the CXCR4 surface signal (detected by mAb 44717) was measured by flow cytometry (n = 3). (B) Antibody-feeding experiments where HL60 cells were first stained with anti-CXCR4 mAb 12G5 and subsequently incubated with the Rac1 inhibitory peptide for different time periods. mAb signal was then measured by flow cytometry (n = 3). (C) Antibody-feeding experiments for which HL60 cells were first stained with anti-CXCR4 mAb 44717 and subsequently incubated with the Rac1 inhibitory peptide for 30 minutes or NSC23766 for 1 hour. mAb signal was subsequently measured by flow cytometry (n = 3). (D) Antibody-feeding experiments for which HL60 cells were first stained with the conformation-independent anti-CXCR4 mAb 4G10 and subsequently incubated with the Rac1 inhibitory peptide for 30 minutes or NSC23766 for 1 hour. mAb signal was subsequently measured by flow cytometry (n = 3). (E) HL60 cells were treated with NSC23766 and CXCR4 surface signal (detected by mAb 44717 or 4G10) was measured (n = 3). Bars show the mean fluorescence intensity determined by flow cytometry and expressed as percentage ± SEM compared with untreated or control conditions (*P < .05, **P < .01).
Rac1 inhibition alters CXCR4 conformation. (A) HL60 cells were incubated with CXCL12 or the Rac1 inhibitory peptide at 4°C or 37°C and the CXCR4 surface signal (detected by mAb 44717) was measured by flow cytometry (n = 3). (B) Antibody-feeding experiments where HL60 cells were first stained with anti-CXCR4 mAb 12G5 and subsequently incubated with the Rac1 inhibitory peptide for different time periods. mAb signal was then measured by flow cytometry (n = 3). (C) Antibody-feeding experiments for which HL60 cells were first stained with anti-CXCR4 mAb 44717 and subsequently incubated with the Rac1 inhibitory peptide for 30 minutes or NSC23766 for 1 hour. mAb signal was subsequently measured by flow cytometry (n = 3). (D) Antibody-feeding experiments for which HL60 cells were first stained with the conformation-independent anti-CXCR4 mAb 4G10 and subsequently incubated with the Rac1 inhibitory peptide for 30 minutes or NSC23766 for 1 hour. mAb signal was subsequently measured by flow cytometry (n = 3). (E) HL60 cells were treated with NSC23766 and CXCR4 surface signal (detected by mAb 44717 or 4G10) was measured (n = 3). Bars show the mean fluorescence intensity determined by flow cytometry and expressed as percentage ± SEM compared with untreated or control conditions (*P < .05, **P < .01).
To examine whether Rac1 controls CXCR4 trafficking, antibody-feeding experiments were performed. Fluorescently labeled anti-CXCR4 mAb (clone 12G5 or 44717) were first allowed to bind to the receptor before addition of the Rac1 inhibitor to the cells. Using flow cytometry we found that inhibition of Rac1 for various time periods resulted in a time-dependent decrease of the fluorescence intensity of the 12G5 CXCR4 antibody signal (Figure 3B). In addition, incubation with the Rac1 inhibitory peptide for 30 minutes or NSC23766 for 1 hour also caused a major decrease in fluorescence intensity of the 44717 CXCR4 antibody signal compared with untreated or control peptide treated cells (Figure 3C). These results were obtained at 37°C, but also at 4°C (data not shown), thereby excluding the internalization of the receptor-antibody complex. These data show that the bound antibodies dissociate on Rac1 inhibition, which is indicative for a conformational change of CXCR4 induced by Rac1 inhibition.
To substantiate this notion, we used the amino-terminus specific CXCR4 mAb, 4G10,21 which is a conformation-independent antibody. Interestingly, the conformation-independent CXCR4 antibody did not dissociate from its receptor after Rac1 inhibition in an antibody-feeding experiment (Figure 3D). This is in contrast to the dissociation of the 12G5 and 44717 antibodies (Figure 3B-C), that have previously been described to be conformation-dependent antibodies.12 In addition, pretreatment of HL60 cells (Figure 3E) or CD34+ (supplemental Figure 1) with NSC23766 did not result in a loss of CXCR4 surface signal measured by 4G10 compared with antibody clones 12G5 and 44717. Our observations that the binding of 4G10 to CXCR4 is not affected by Rac1 inhibition, confirm that the receptor is not internalized, but rather adopts a new conformation on Rac1 inhibition.
CXCR4 conformational change on Rac1 inhibition abrogates receptor internalization and impairs Gαi signaling
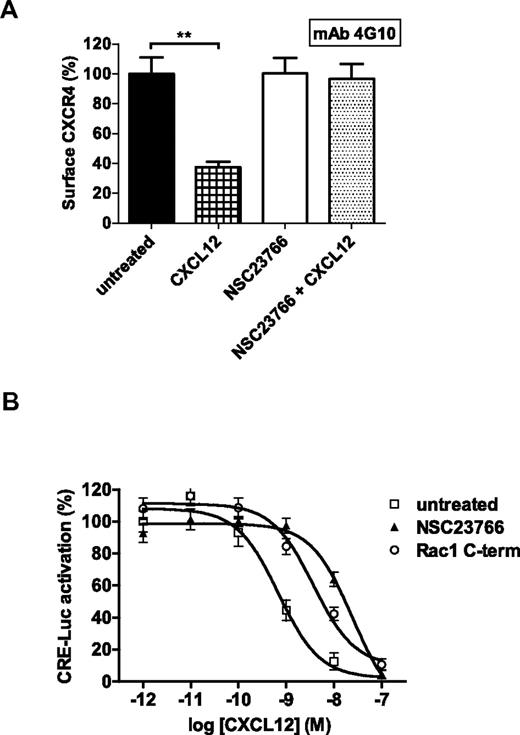
To study the functional consequences of the CXCR4 conformational change, we measured ligand-induced receptor endocytosis after Rac1 inhibition. To this end, we used the conformation-independent antibody clone, 4G10. In Jurkat T cells, CXCL12 stimulation induced a significant decrease of CXCR4 surface expression, which is indicative for receptor endocytosis. However, after NSC23766 treatment, ligand stimulation did not cause receptor internalization suggesting that the conformational change in CXCR4 on inhibition of Rac1 blocks CXCL12 binding (Figure 4A).
CXCR4 internalization and Gαi signaling are impaired after CXCR4 conformational change on Rac1 inhibition. (A) Jurkat T cells were pretreated with or without NSC23766 and subsequently incubated with or without CXCL12. The CXCR4 (detected by mAb 4G10) surface signal was then measured (n = 3). Bars show the mean fluorescence intensity determined by flow cytometry and expressed as percentage ± SEM compared with untreated conditions. (B) HEK 293T cells expressing CXCR4 and CRE-Luc were stimulated with forskolin and different concentrations of CXCL12 (varied from 1pM to 100nM) in the absence or presence of NSC23766 or the Rac1 inhibitory peptide. After 8 hours, CXCL12-mediated inhibition of cyclic AMP synthesis was analyzed by the luciferase assay as indicated in “CRE-luciferose reporter gene assay.” The graph shows normalized data of 3 independent experiments where 100% represents the luciferase activity of untreated cells at the lowest CXCL12 concentration (**P < .01).
CXCR4 internalization and Gαi signaling are impaired after CXCR4 conformational change on Rac1 inhibition. (A) Jurkat T cells were pretreated with or without NSC23766 and subsequently incubated with or without CXCL12. The CXCR4 (detected by mAb 4G10) surface signal was then measured (n = 3). Bars show the mean fluorescence intensity determined by flow cytometry and expressed as percentage ± SEM compared with untreated conditions. (B) HEK 293T cells expressing CXCR4 and CRE-Luc were stimulated with forskolin and different concentrations of CXCL12 (varied from 1pM to 100nM) in the absence or presence of NSC23766 or the Rac1 inhibitory peptide. After 8 hours, CXCL12-mediated inhibition of cyclic AMP synthesis was analyzed by the luciferase assay as indicated in “CRE-luciferose reporter gene assay.” The graph shows normalized data of 3 independent experiments where 100% represents the luciferase activity of untreated cells at the lowest CXCL12 concentration (**P < .01).
To determine CXCR4 signaling efficiency on Rac1 inhibition in Jurkat T cells, we investigated the capacity of CXCR4 to inhibit cAMP production as a result of Gαi protein activation. Using a cAMP response element (CRE) reporter gene in transiently transfected HEK293T cells, CXCL12 stimulation inhibited the forskolin-induced CRE activation (pEC50 = 9.1 ± 0.1, n = 3). Inhibition of Rac1 drastically reduced CXCL12-mediated cAMP inhibition as illustrated by a significant shift to the right of the CXCL12 concentration response curve (pEC50 for NSC23766 = 7.7 ± 0.1; (P < .01) and pEC50 for the Rac1 inhibitory peptide = 8.7 ± 0.1, (P < .05), n = 3; Figure 4B). This means that the conformation of CXCR4 adopted after Rac1 inhibition has a decreased capacity in initiating Gαi protein signaling. These data suggest that CXCR4 becomes less sensitive to CXCL12 on Rac1 inhibition. Overall, the lack of receptor internalization and impaired Gαi signaling after Rac1 inhibition demonstrate that Rac1 activity is required for the maintenance of the responsive CXCR4 conformation.
HIV-1 infection is inhibited after CXCR4 conformational change
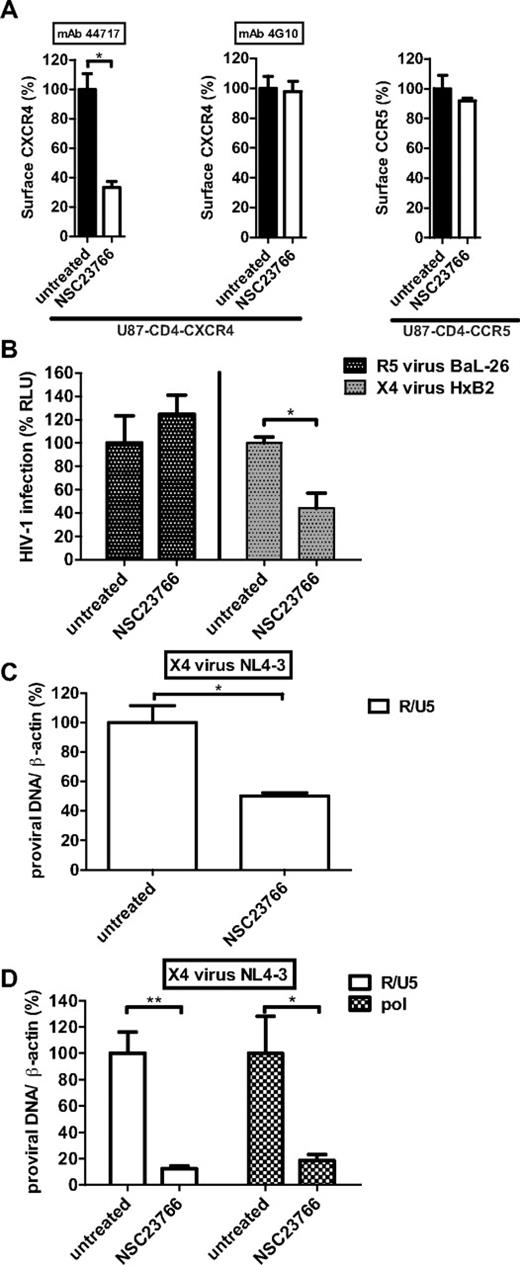
Two chemokine receptors CXCR4 and CCR5 have been identified as coreceptors for HIV-1, and viral tropism is defined by the use of either CXCR4 (X4 viruses) or CCR5 (R5 viruses).4,22 To evaluate whether the CXCR4 conformational change caused by Rac1 inhibition affects HIV-1 binding and infection, we initially performed an infection assay using a single-round, luciferase reporter HIV-1 (NL4-3.Luc.R-E-) pseudotyped with a CXCR4-using envelope (HxB2) or a CCR5-using envelope (BaL-26). U87 cells stably expressing CD4 in combination with CXCR4 or CCR5 were used as host cells in this assay. Treating these cells with NSC23766 caused a conformational change of CXCR4, as determined by the loss of conformation-dependent antibody binding without affecting the binding of the conformation-independent 4G10 antibody clone (Figure 5A left and middle panels). The conformation of CCR5 was unaffected by NSC23766 treatment (Figure 5A right panel). As shown in Figure 5B, NSC23766 treatment markedly reduced infection of luciferase reporter HIV-1 pseudotyped with the X4-using HxB2 envelope compared with untreated cells. Several reports identified Rac1 and its downstream effectors, involved in actin dynamics, as crucial players in the entry of R5 viruses into host cells.23,24 However, in our system, R5 HIV-1 infection was independent of Rac1, because NSC23766 treatment did not affect the infection of luciferase reporter HIV-1 pseudotyped with the R5-using BaL-26. These data confirm the specificity of Rac1 for regulating CXCR4 conformation without interfering with CCR5.
HIV-1 infection is blocked after CXCR4 conformational change. (A) U87-CD4-CXCR4 and U87-CD4-CCR5 cells were incubated with NSC23766 and CXCR4 (detected by mAb 44717 or 4G10) and CCR5 (detected by mAb 2D7) surface signals were measured. Bars show the mean fluorescence intensity determined by flow cytometry and expressed as percentage ± SEM compared with untreated conditions (n = 3). (B) U87-CD4-CXCR4, and U87-CD4-CCR5 cells were inoculated, in the presence or absence of NSC23766, with the single round luciferase reporter HIV-1 pseudotyped with the X4-using envelope HxB2 or the R5-using envelope BaL-26, respectively. Forty-eight hours later, the infectivity of the virus was analyzed by the luciferase assay. Bars show the luciferase activity measured by a microplate luminometer and expressed as percentage ± SEM compared with untreated conditions (n = 3). (C) U87-CD4-CXCR4 cells were inoculated with the X4-virus NL4-3 in the presence or absence of NSC23766. After 48 hours, DNA samples were isolated and the early reverse transcriptase product R/U5 was quantified by qPCR. Bars show the number of proviral DNA copies corrected for differences in DNA input (β-actin) and expressed as percentage ± SEM compared with untreated conditions (n = 3). (D) PHA-stimulated PBMCs were inoculated with the X4-virus NL4-3 in the presence or absence of NSC23766. After 48 hours, DNA samples were isolated and R/U5 and pol proviral DNA were quantified by qPCR. Bars show the number of proviral DNA copies corrected for differences in DNA input (β-actin) and expressed as percentage ± SEM compared with untreated conditions (n = 4; *P < .05, **P < .01).
HIV-1 infection is blocked after CXCR4 conformational change. (A) U87-CD4-CXCR4 and U87-CD4-CCR5 cells were incubated with NSC23766 and CXCR4 (detected by mAb 44717 or 4G10) and CCR5 (detected by mAb 2D7) surface signals were measured. Bars show the mean fluorescence intensity determined by flow cytometry and expressed as percentage ± SEM compared with untreated conditions (n = 3). (B) U87-CD4-CXCR4, and U87-CD4-CCR5 cells were inoculated, in the presence or absence of NSC23766, with the single round luciferase reporter HIV-1 pseudotyped with the X4-using envelope HxB2 or the R5-using envelope BaL-26, respectively. Forty-eight hours later, the infectivity of the virus was analyzed by the luciferase assay. Bars show the luciferase activity measured by a microplate luminometer and expressed as percentage ± SEM compared with untreated conditions (n = 3). (C) U87-CD4-CXCR4 cells were inoculated with the X4-virus NL4-3 in the presence or absence of NSC23766. After 48 hours, DNA samples were isolated and the early reverse transcriptase product R/U5 was quantified by qPCR. Bars show the number of proviral DNA copies corrected for differences in DNA input (β-actin) and expressed as percentage ± SEM compared with untreated conditions (n = 3). (D) PHA-stimulated PBMCs were inoculated with the X4-virus NL4-3 in the presence or absence of NSC23766. After 48 hours, DNA samples were isolated and R/U5 and pol proviral DNA were quantified by qPCR. Bars show the number of proviral DNA copies corrected for differences in DNA input (β-actin) and expressed as percentage ± SEM compared with untreated conditions (n = 4; *P < .05, **P < .01).
To further support that the infection of X4-using HIV-1 was inhibited at the level of virus entry, additional infection experiments were performed analyzing early steps after virus entry by qPCR. U87-CD4-CXCR4 cells were inoculated with a replicating X4-virus (NL4-3) and 48 hours later total DNA was isolated and analyzed for the presence of early (R/U5) products of reverse transcription. In agreement with the data from the HIV-1 luciferase assay, the detected levels of R/U5 were significantly lower in NSC23766-treated cells compared with untreated conditions (Figure 5C). These results were further confirmed in X4-using HIV-1 infection experiments using PHA-stimulated PBMCs. Figure 5D shows that only low levels of R/U5 and the relatively late (pol) product of reverse transcription were detected in NSC23766-treated PBMCs compared with untreated cells. As the presence of R/U5 proviral DNA is indicative for efficient virus entry and initiation of reverse transcription, these data demonstrate that Rac1 inhibition impairs virus entry by altering CXCR4 conformation. These data correlate with the lack of CXCR4 internalization after Rac1 inhibition as observed in Figure 4A.
Discussion
Although the conformational change of CXCR4 that follows ligand binding is established,5 less is known about the ligand-independent regulation of CXCR4 conformation in resting cells. Here, we show that the conformation of CXCR4 in resting conditions is of major importance as it defines the responsiveness to the CXCL12 chemotactic stimulus and susceptibility for HIV-1 binding and entry into host cells. The link between CXCR4 and Rac1 was previously established by showing that Rac1 is an important downstream signaling molecule translating the CXCL12 stimulus to cytoskeletal remodeling and cell movement.25 Interestingly, we found that Rac1 associates with CXCR4 via the Rac1 C-terminus independently of GDP-GTP loading of the GTPase or ligand stimulation. Furthermore, Rac1 controls CXCR4 conformation specifically, because Rac1 inhibition did not affect the conformation of the related chemokine receptors CCR5 and CXCR7. Finally, Rac1 regulation of CXCR4 conformation was not cell-type specific, because we observed similar results in different cell types, including CD34+ HSPCs. Thus, our findings identify Rac1 as a positive regulator of CXCR4.
Furthermore, the lack of receptor internalization and reduced capacity to activate Gαi protein after Rac1 inhibition, demonstrates that Rac1 activity is required for the maintenance of the responsive CXCR4 conformation. This suggests that Rac1 acts as an intracellular positive allosteric modulator of CXCR4, which stabilizes the responsive conformation of the receptor.
Baribaud et al described the presence of at least 2 antigenically distinct conformations of CXCR4 in resting conditions.12 This heterogeneity in CXCR4 populations was not because of differences in N-linked glycosylation or sulfation of the receptor. Our preliminary results identified the third intracellular loop of CXCR4 as an important domain for defining the receptor's conformation (Y.Z., unpublished data, May 1, 2011). Interestingly, we have previously shown that by modifying intracellular parts of the viral chemokine receptor ORF74, the conformation of the receptor and binding of its ligand(s) is altered.26 Further work will unravel whether Rac1 and the third intracellular loop of CXCR4 act in concordance to regulate the conformation adopted by this receptor.
It is not clear how Rac1 maintains CXCR4 in the responsive conformation. It has been shown that disruption of cholesterol-rich lipid raft domains causes a conformational change of CXCR4.27 Active Rac1 is localized in lipid raft microdomains28,29 and may preserve a pool of CXCR4 in a responsive conformation at these specific signaling platforms on the cell membrane. These CXCR4 pools would represent the receptor subset with high signal-transduction capacity and HIV-1 coreceptor activity.30,31
The fact that Rac1 inhibition impaired the binding of the 2 tested conformational-dependent anti-CXCR4 antibodies, which both recognize an epitope in the second extracellular loop of CXCR4, implies that at least the conformation of this domain is altered on Rac1 inhibition. Moreover, the second extracellular loop plays a prominent role in HIV-1 gp120 recognition and binding.14,32 In line with this, we found that CXCR4 conformation after Rac1 inhibition prevented CXCR4-dependent HIV-1 infection. Thus, next to a specific domain in the coreceptor, viral entry is also dependent on the conformation adopted by the coreceptor.
Our data, which show that Rac1 inhibition alters the conformation of CXCR4 expressed on CD34+ HSPCs, may also have important clinical implications. First, Carter et al33 recently showed that CXCR4-tropic HIV-1 can infect HSCs thereby creating a latent viral reservoir which could be responsible for the disease progression of AIDS. Therefore, manipulating CXCR4 conformation on this cell population will contribute to the eradication of the latent HIV-1 reservoir. Next, a change of CXCR4 conformation, which no longer supports its binding to CXCL12, could improve HSC mobilization because the retention of these cells in the bone marrow depends on the CXCR4-CXCL12 axis. In line with this assumption, it has been shown that intraperitoneal administration of NSC23766 in mice caused a massive mobilization of HSCs into the circulation.34 A change of CXCR4 conformation after NSC23766 administration, which breaks the CXCR4-CXCL12 interaction, is a possible mechanism explaining the tremendous egress of HSCs from the bone marrow.
In conclusion, we established Rac1 as a key player in the regulation of CXCR4 conformation, thereby controlling the fine-tuning of CXCR4 activity and affecting HIV-1 binding and fusion. Finally, uncovering the molecular mechanisms governed by Rac1 and directed at the regulation of CXCR4 conformation, will provide novel strategies to block infection of X4 viruses, which have previously been associated with an increased progression rate to AIDS.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Marieke von Lindern (Department of Hematopoiesis, Sanquin Research and Landsteiner Laboratory, Academic Medical Center, University of Amsterdam, Amsterdam, The Netherlands) for helpful discussion and comments, and the Lansteiner Foundation for Blood Transfusion Research (LSBR) for funding (grant no. 06.30).
Authorship
Contribution: Y.Z., K.B., D.M., and K.A.v.D. performed experiments; Y.Z., C.V., N.A.K, M.J.S., D.M., P.L.H., and P.B.v.H. designed the research and analyzed the results; and Y.Z. and P.B.v.H made the figures and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Paula B. van Hennik, Sanquin Research, Dept of Hematopoiesis, Plesmanlaan 125, 1066 CX Amsterdam, The Netherlands; e-mail: p.vanhennik@sanquin.nl.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal