Abstract
The t(8;21) RUNX1-ETO translocation is one of the most frequent cytogenetic abnormalities in acute myeloid leukemia (AML). In RUNX1-ETO+ patient samples, differing classes of activating c-KIT receptor tyrosine kinase mutations have been observed. The most common (12%-48%) involves mutations, such as D816V, which occur in the tyrosine kinase domain, whereas another involves mutations within exon 8 in a region mediating receptor dimerization (2%-13% of cases). To test whether distinct subtypes of activating c-KIT mutations differ in their leukemogenic potential in association with RUNX1-ETO, we used a retroviral transduction/transplantation model to coexpress RUNX1-ETO with either c-KitD814V or c-KitT417IΔ418-419 in murine hematopoietic stem/progenitor cells used to reconstitute lethally irradiated mice. Analysis of reconstituted animals showed that RUNX1-ETO;c-KitD814V coexpression resulted in 3 nonoverlapping phenotypes. In 45% of animals, a transplantable AML of relatively short latency and frequent granulocytic sarcoma was noted. Other mice exhibited a rapidly fatal myeloproliferative phenotype (35%) or a lethal, short-latency pre-B-cell leukemia (20%). In contrast, RUNX1-ETO;c-KitT417IΔ418-419 coexpression promoted exclusively AML in a fraction (51%) of reconstituted mice. These observations indicate that c-KitD814V promotes a more varied and aggressive leukemic phenotype than c-KitT417IΔ418-419, which may be the result of differing potencies of the activating c-Kit alleles.
Introduction
Chromosomal translocations involving genes encoding the 2 subunits of the heterodimeric core-binding factor (CBF) transcription factor complex, RUNX1 and CBFβ, are the most frequent cytogenetic abnormalities in acute myeloid leukemia (AML).1,2 The t(8;21)(q22;q22) rearrangement results in an in-frame fusion of the N-terminal 177 amino acids of RUNX1, including the DNA-binding Runt domain, to nearly the entire ETO protein generating RUNX1-ETO (RE).3,4 ETO (also known as RUNX1T1, or MTG8) can interact with the nuclear corepressors N-CoR, mSin3, SMRT, and various histone deacetylases,5-8 so it may function in RE by altering the normal expression pattern of CBF target genes. Observations showing that RUNX1-ETO heterozygous knock-in mice phenocopy RUNX1 homozygous knockout mice support the view that RE functions as a dominant inhibitor of normal CBF activity.9-12 The t(8;21) is thought to be an early event in AML pathogenesis as the translocation can be detected at relatively high frequencies in utero.13 Murine studies have also shown that RE promotes gradual but significant accumulation of myeloid progenitor cells that have enhanced serial replating potential in vitro and some capacity for myeloid differentiation.9,14-16 In these studies, mice rarely progress to AML. The relatively weak oncogenic potential of RE is consistent with observations showing that cells from patients in long-term clinical remission continue to express RUNX1-ETO transcripts.17
A number of studies have defined cooperating mutations that significantly accelerate progression to AML in association with RUNX1-ETO,18 including treatment of RE+ mice with ENU15 and coexpression of RE with TEL-PDGFβR,19 FLT3-ITD,20 and Wilms tumor 1.21 Acceleration of AML was also noted when RE was expressed in ICSBP22 or p2123 knockout BM cells. In up to 70% of human t(8;21)+ patient samples, nonoverlapping mutations in receptor tyrosine kinases (RTKs), particularly c-KIT, but also FLT3 to a lesser degree, or mutations in NRAS have been noted.24-33 c-KIT encodes a type III RTK, characterized by 5 immunoglobulin-like repeats in the extracellular portion of the molecule, a transmembrane domain, and a cytoplasmic region that includes a juxtamembrane domain encoded by exon 11 and a kinase domain split by an insert into an ATP-binding region and a phosphotransferase domain encoded by exon 17.34,35 Although c-KIT–activating mutations have only been observed in 2% to 5% of total AML cases, they are disproportionately present in 12% to 48% of t(8;21)+ patient samples.26,30 The most common class of c-KIT mutation in t(8;21)+–associated AML occurs in the activation loop of the kinase domain, resulting in D816V (D814V in mice) or N822K in 11% to 44% of cases. A second class, occurring in 2% to 13% of t(8;21)+ samples, involves mutations within exon 8 in the extracellular portion of the receptor that encodes the c-KIT dimerization domain.18,25-27,29,30,32,36 The D816V and N822K mutations in the activation loop may not be functionally analogous given that each is differentially sensitive to the tyrosine kinase inhibitor imatinib.30,37 Exon 8 mutations leading to hyperactivation of the receptor are heterogeneous and involve small deletions/insertions affecting amino acids 417 to 419. One well-characterized exon 8 mutant (c-KitT417IΔ418-419) observed in patients replaces a threonine residue at amino acid 417 with isoleucine and includes a small deletion of amino acids 418 to 419.25,38 The ability of exon 8 mutations to promote progression to AML in association with RUNX1-ETO in vivo has not been explored.
In murine studies, retroviral expression of c-KitD814V in a number of hematopoietic progenitor cell lines resulted in factor-independent growth in vitro and leukemia within 6 to 19 weeks in 6 of 10 transplanted animals. Leukemic cells were largely positive for B220 and lacked expression of Gr-1 and Mac-1, suggesting that the leukemias were largely immature pre-B cell acute lymphocytic leukemia (B-ALL).39 Transgenic mice expressing c-KitD814V from an MHC class I H-2Ld promoter similarly developed B-ALL, T-cell acute lymphocytic lymphoma (T-ALL), or lymphomas in 4 of 15 transgenic animals.39 In another study, retroviral expression of either a chimeric c-Kit molecule fusing the extracellular and transmembrane portions of murine c-Kit in-frame with the intracellular signaling domain of human c-KITD816V resulted in rapidly lethal myeloproliferative disease (MPD; median latency of 40-70 days) characterized by leukocytosis, splenomegaly, and infiltration of Mac-1+ cells in peripheral tissues in 100% of reconstituted mice.40 Finally, a very recent study using a retroviral transduction/transplantation approach showed that retroviral expression of the c-KitN822K activation loop mutant resulted in lethal MPD with a median survival of 171 days.37 Mice reconstituted with cells that only expressed a c-Kit juxtamembrane mutant developed a rapidly fatal B220+CD19+ B-ALL in two-thirds of transplanted mice or a fatal MPD in the remaining one-third of mice. Coexpression of RUNX1-ETO with c-KitN822K resulted in lethal AML in approximately 70% of transplanted mice with a median latency of 177 days, which was similar to the latency observed in mice transplanted with c-KitN822K–expressing cells.37
In this study, we used a retroviral transduction/transplantation approach to compare the relative ability of an activating c-Kit extracellular domain mutation (c-KitT417IΔ418-419) versus a c-Kit mutation in the activation loop (c-KitD814V) to cooperate with RE in promotion of AML. Analysis of reconstituted mice showed that RE;c-KitD814V coexpression resulted in 3 nonoverlapping phenotypes, including AML (45%), a myeloproliferative neoplasm (MPN; 35%), and pre-B-ALL (20%). RE changed both the latency and the neoplastic phenotype of mice compared with animals that only expressed c-KitD814V, which predominantly developed lethal MPD or T-ALL. In contrast, RE;c-KitT417IΔ418-419 mice developed exclusively AML in a fraction (51%) of reconstituted animals with a median latency that was nearly double that observed in RE;c-KitD814V mice that developed AML. Analysis of clonality showed that additional genetic changes were likely necessary for leukemic progression in all reconstituted mice, suggesting that these 2 “hits” were not sufficient for AML. Differences in neoplastic phenotype between RE;c-KitD814V and RE;c-KitT417IΔ418-419 mice indicate that different classes of c-Kit–activating mutations differ in their ability to promote AML, which may account for the observed differences in prevalence of distinct c-Kit–activating mutations associated with t(8;21)+ leukemia.
Methods
Cloning
Wild-type murine c-Kit cDNA was PCR-amplified from pEF-BOS-Wt-c-Kit (kindly provided by Dr Itaru Matsumura, Osaka, Japan) and cloned into MSCV-IRES-Vex (MIV)41 generating MSCV-c-KitWt-IRES-Vex. Retroviral vectors expressing c-KitD814V or c-KitT417IΔ418-419 were made by site-directed mutagenesis of MSCV-c-KitWt-IRES-Vex using QuikChange II XL (Agilent Technologies). The MSCV-IRES-Bex (MIB) and MSCV-RUNX1-ETO-IRES-Bex (MIB-RE) vectors have been described.16
Retroviral transduction and transplantation assays
Transductions were performed according to Cotta et al42 (supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Animal research was performed in accordance with the Institutional Animal Care and Use Committee at University of Alabama at Birmingham under the supervision of the Association For Laboratory Animal Care (ALAC) accredited veterinary staff.
Flow cytometric analysis of cell-surface and intracellular c-Kit expression
NIH 3T3 cells were transduced with MIV, MIV-c-KitWt, MIV-c-KitD814V, or MIV-c-KitT417Δ418-419 in the presence of 5 μg/mL polybrene for 24 hours. Transduced cells were stained with anti–c-Kitbiotin (2B8), followed by streptavidinFITC for detection of membrane c-Kit. Intracellular FACS was performed using BrdU Flow Kit and anti–c-KitAPC (2B8) according to the manufacturer's protocol (BD Biosciences). Flow cytometry was performed using a 3-laser LSRII (BD Biosciences) and data analyzed using FlowJo Version 6.4.7 software (TreeStar).
BM, PB, spleen, and sarcoma flow cytometry and cell sorting
Peripheral blood (PB) and BM harvest were performed as described.42 Splenocytes and granulocytic sarcoma cells were obtained by mechanical disruption and resuspended in HBSS/2% FBS. Cells were stained with: anti–Mac-1APC (M1/70), anti–Gr-1PE-Cy7 (RB6-8C5), anti-CD115PE (AFS98), anti-B220eFluor450 (RA3-6B2), anti-CD19PE-Cy7 (1D3), anti-CD43PE (S7), anti-IgMAPC (RMM-1; BioLegend), anti-CD5PE (53-7.3) anti–CD4PE or APC (GK1.5 or RM4-5), anti–CD8PE or PE-Cy7 (53-6.7), anti-CD25PE (PC61), and anti–CD44PE-Cy5 or APC (IM7). Sorting was performed using a 3-laser MoFlo (Dako Cytomation) to purity > 95%. Dead cells were excluded by propidium iodide. Antibodies were purchased from eBioscience or BD Biosciences unless noted.
Microscopy
Images were acquired using a Zeiss Axio Imager A1 microscope (Carl Zeiss) equipped with a Zeiss AxioCam MRc camera and Zeiss AxioVision Version 4.5 software. Zeiss EC-Plan-NEOFLUAR 10×/0.3 and Zeiss Plan-APOCHROMAT 63×/1.4 oil objectives were used for image capture.
Colony-forming assays
Bex+Vex+ myeloid scatter-gated BM cells were FACS-purified into DMEM/10% FBS and 1 × 104 cells plated in duplicate in methylcellulose media containing SCF, IL-3, IL-6, and erythropoietin (M3434; StemCell Technologies). Total numbers of myeloid colony-forming units (CFU) per plate were counted after 7 to 12 days of culture in 5% CO2 at 37°C.
Southern blot
Genomic DNA was prepared from whole splenocytes of moribund RE;c-KitD814V or RE;c-KitT417IΔ418-419 animals. A total of 10 μg of each genomic DNA sample was digested with NcoI, separated on a 0.75% agarose gel, and then transferred to Hybond XL nylon membranes (GE Healthcare). Blots were hybridized with α-32P[dCTP]–labeled probes complementary to c-Kit or ETO sequences. Probes were radioactively labeled with Rediprime II Random Prime Labeling System (GE Healthcare). Retroviral vectors contained a single NcoI site localized downstream of the probe-binding region, allowing detection of unique proviral integrants. Genomic DNA isolated from splenocytes of wild-type C57BL/6-Ly5.1 mice served as controls for detection of endogenous sequences.
Results
Differing potencies of c-KitD814V or c-KitT417IΔ418-419 to promote AML when coexpressed with RUNX1-ETO
Because distinct subtypes of activating mutations in c-KIT occur at different frequencies in t(8;21)+ patient samples, we wished to test whether this might impact their ability to cooperate with RE in promotion of AML. To address this, we cotransduced an enriched population of hematopoietic stem/progenitor cells with 2 MSCV-based retroviral vectors, with one construct coexpressing RE and a blue-excited GFP variant (Bex) and the other retrovirus coexpressing either c-KitD814V or c-KitT417IΔ418-419 in conjunction with a violet-excitable GFP reporter, Vex (Figure 1A).41 c-Kit–activating mutations were verified by DNA sequencing, and protein expression was confirmed by FACS analysis and Western blotting (Figure 1B; and data not shown). These results showed that Vex protein levels correlated with levels of coexpressed c-Kit mutants and that c-KitD814V was expressed at much lower levels on the cell surface than wild-type c-Kit or the c-KitT417IΔ418-419 mutant, even though it was expressed in the cytoplasm at significant levels. These results confirm previous studies showing that, in contrast to wild-type c-Kit, c-KitD814V is expressed at lower levels on the plasma membrane but is abundantly expressed in the Golgi, where it maintains its signaling and transforming capabilities.37,40,43 Transduced cells were transplanted into lethally irradiated, C57BL/6-Ly5.2 congenic recipient mice, which were then monitored for hematopoietic defects and leukemia development in both single- and double-transduced cells beginning at 3 weeks post-transplant.
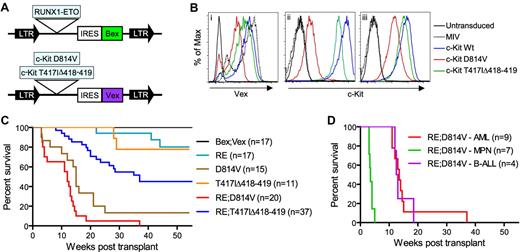
Generation of animals transplanted with RE;c-KitD814V– or RE;c-KitT417IΔ418-419–expressing cells. (A) Representation of MSCV-based retroviral constructs. LTR indicates long-terminal repeats; and IRES, internal ribosome entry site. (B) FACS analysis of NIH 3T3 cells transduced with MIV, c-KitWt, c-KitD814V, or c-KitT417IΔ418-419 retroviruses, demonstrating Vex expression (i), surface c-Kit (ii), and intracellular c-Kit (iii) levels. (C) Kaplan-Meier survival analysis of mice transplanted with BM cells expressing control Bex;Vex (n = 17), RUNX1-ETO (RE; n = 17), D814V (n = 15), T417IΔ418-419 (n = 11), RE;c-KitD814V (n = 20), or RE;c-KitT417IΔ418-419 (n = 37) retroviruses. (D) Kaplan-Meier survival analysis depicting differing latencies of RE;c-KitD814V–associated neoplasia, which included AML (n = 9), MPN (n = 7), and pre-B-ALL phenotypes (n = 4).
Generation of animals transplanted with RE;c-KitD814V– or RE;c-KitT417IΔ418-419–expressing cells. (A) Representation of MSCV-based retroviral constructs. LTR indicates long-terminal repeats; and IRES, internal ribosome entry site. (B) FACS analysis of NIH 3T3 cells transduced with MIV, c-KitWt, c-KitD814V, or c-KitT417IΔ418-419 retroviruses, demonstrating Vex expression (i), surface c-Kit (ii), and intracellular c-Kit (iii) levels. (C) Kaplan-Meier survival analysis of mice transplanted with BM cells expressing control Bex;Vex (n = 17), RUNX1-ETO (RE; n = 17), D814V (n = 15), T417IΔ418-419 (n = 11), RE;c-KitD814V (n = 20), or RE;c-KitT417IΔ418-419 (n = 37) retroviruses. (D) Kaplan-Meier survival analysis depicting differing latencies of RE;c-KitD814V–associated neoplasia, which included AML (n = 9), MPN (n = 7), and pre-B-ALL phenotypes (n = 4).
We observed that all mice coexpressing RE and c-KitD814V (RE;c-KitD814V mice) developed lethal hematopoietic neoplasms of diverse phenotype between 2 and 4 months post-transplant that included AML (45%), an MPN (35%), and pre-B ALL in 20% of animals (Figure 1C; Table 1). Moribund RE;c-KitD814V mice presenting with AML or pre-B cell ALL progressed with similar kinetics at 3 to 4 months after transplantation, whereas mice with MPN died within 3 to 5 weeks of transplantation (Figure 1D). Animals reconstituted with cells only expressing RE (n = 17) rarely progressed to lethal disease within 1 year post-transplant, as noted previously (Figure 1C).14-16,44 However, ∼ 90% of mice reconstituted with cells only expressing c-KitD814V died from either MPN in half of the moribund mice or ALL that was predominantly T-ALL (Figure 1C; Table 1). RE;c-KitD814V coexpression accelerated the onset of death by approximately 1 month compared with c-KitD814V mice (Figure 1C) and significantly altered the type of hematologic malignancy that developed since AML was the predominant phenotype in RE;c-KitD814V mice (Table 1). AML was characterized by high frequencies of myeloid blasts that generally represented 60% to 80% of Bex+Vex+ nucleated WBCs in BM of moribund animals (Figure 2A). In contrast to RE;c-KitD814V mice, approximately 50% of animals transplanted with RE;c-KitT417IΔ418-419–expressing cells never developed a lethal hematologic neoplasm within the one year post-transplant that mice were observed (Figure 1C). In addition, AML with a median onset of approximately 4 to 5 months was the only phenotype noted among the 50% of RE;c-KitT417IΔ418-419 mice that became moribund, although there were discernable differences in the frequencies of differentiated myeloid cells between moribund RE;c-KitT417IΔ418-419 animals (Figure 3B). RE;c-KitT417IΔ418-419 mice could display high levels of Bex+Vex+ expression for long periods of time (months) before any significant changes in chimerism indicative of additional genetic changes leading to clonal expansion were observed (note differences between 2 subsets of Bex+Vex+ cells in mouse A, supplemental Figure 1).
Hematopoietic neoplasia in reconstituted mice
| Retroviral construct(s) . | No. of mice . | AML (sarcoma) . | MPN . | B-ALL . | T-ALL . |
|---|---|---|---|---|---|
| Bex;Vex | 17 | 0 | 0 | 0 | 0 |
| RE | 17 | 3 (ND) | 0 | 0 | 0 |
| D814V | 15 | 0 | 7 | 1 | 5 |
| T417IΔ418-419 | 11 | 0 | 0 | 2 | 0 |
| RE;T417IΔ418-419 | 37 | 19 (3) | 0 | 0 | 0 |
| RE;D814V | 20 | 9 (7) | 7 | 4 | 0 |
| Retroviral construct(s) . | No. of mice . | AML (sarcoma) . | MPN . | B-ALL . | T-ALL . |
|---|---|---|---|---|---|
| Bex;Vex | 17 | 0 | 0 | 0 | 0 |
| RE | 17 | 3 (ND) | 0 | 0 | 0 |
| D814V | 15 | 0 | 7 | 1 | 5 |
| T417IΔ418-419 | 11 | 0 | 0 | 2 | 0 |
| RE;T417IΔ418-419 | 37 | 19 (3) | 0 | 0 | 0 |
| RE;D814V | 20 | 9 (7) | 7 | 4 | 0 |
ND indicates not determined.
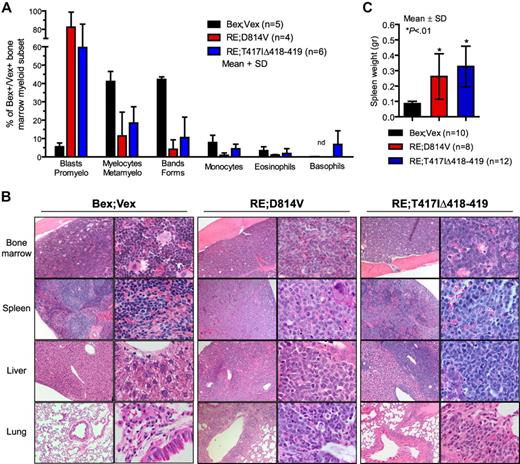
AML blasts in BM and peripheral tissues. (A) Differential counts were performed on Wright-Giemsa–stained cytospins of FACS-purified Bex+Vex+ myeloid scatter-gated cells isolated from BM of control Bex;Vex (n = 5), moribund RE;c-KitD814V (n = 4), and moribund RE;c-KitT417IΔ418-419 (n = 6) mice. Data are mean ± SD (percentages) of the indicated myeloid cell subsets determined by typing 350 to 500 cells per sample. nd indicates not detected. (B) Representative H&E-stained tissue sections of BM, spleen, liver, and lung from Bex;Vex control, leukemic RE;c-KitD814V, and leukemic RE;c-KitT417IΔ418-419 mice. Data are representative of a minimum of 5 moribund RE;c-KitD814V and RE;c-KitT417IΔ418-419 mice. Original magnifications in the left and right columns of Bex;Vex, RE;c-KitD814V, and RE;c-KitT417IΔ418-419 images were ×100 and ×630, respectively. (C) Splenomegaly was observed in all moribund RE;c-KitD814V and RE;c-KitT417IΔ418-419 mice. Spleen weights (grams) are presented as mean ± SD for Bex;Vex controls (n = 10), RE;c-KitD814V (n = 8), and RE;c-KitT417IΔ418-419 (n = 12). *P < .01, compared with Bex;Vex controls (unpaired t test).
AML blasts in BM and peripheral tissues. (A) Differential counts were performed on Wright-Giemsa–stained cytospins of FACS-purified Bex+Vex+ myeloid scatter-gated cells isolated from BM of control Bex;Vex (n = 5), moribund RE;c-KitD814V (n = 4), and moribund RE;c-KitT417IΔ418-419 (n = 6) mice. Data are mean ± SD (percentages) of the indicated myeloid cell subsets determined by typing 350 to 500 cells per sample. nd indicates not detected. (B) Representative H&E-stained tissue sections of BM, spleen, liver, and lung from Bex;Vex control, leukemic RE;c-KitD814V, and leukemic RE;c-KitT417IΔ418-419 mice. Data are representative of a minimum of 5 moribund RE;c-KitD814V and RE;c-KitT417IΔ418-419 mice. Original magnifications in the left and right columns of Bex;Vex, RE;c-KitD814V, and RE;c-KitT417IΔ418-419 images were ×100 and ×630, respectively. (C) Splenomegaly was observed in all moribund RE;c-KitD814V and RE;c-KitT417IΔ418-419 mice. Spleen weights (grams) are presented as mean ± SD for Bex;Vex controls (n = 10), RE;c-KitD814V (n = 8), and RE;c-KitT417IΔ418-419 (n = 12). *P < .01, compared with Bex;Vex controls (unpaired t test).
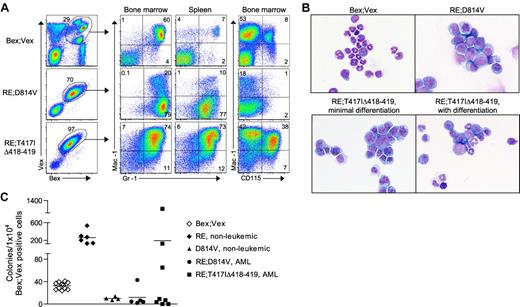
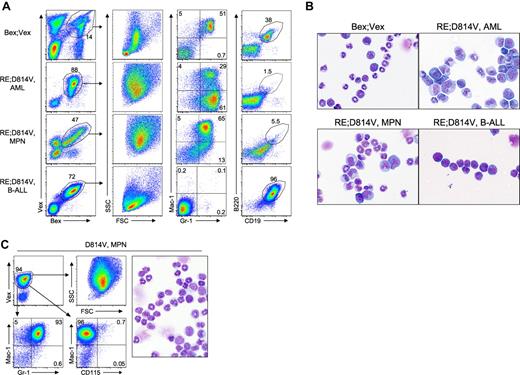
Myeloid-lineage phenotype of leukemic cells in RE;c-KitD814V and RE;c-KitT417IΔ418-419 mice. (A) Representative FACS analysis of AML cells harvested from BM and spleen of Bex;Vex control and moribund RE;c-KitD814V and RE;c-KitT417IΔ418-419 mice. Cells were gated for Bex+Vex+ expression and then analyzed for Mac-1, Gr-1, and CD115 (M-CSFR) expression. (B) Wright-Giemsa–stained cytospins of FACS-purified Bex+Vex+ myeloid-gated BM cells from control or moribund animals demonstrating a high frequency of blast forms in leukemic RE;c-KitD814V and RE;c-KitT417IΔ418-419 mice (original magnification ×630). (C) Myeloid CFU assays performed by plating 1 × 104 FACS-purified Bex+Vex+ myeloid scatter-gated BM cells from Bex;Vex control, nonleukemic RE (39-45 weeks post-transplant), nonleukemic D814V (3-17 weeks post-transplant), moribund RE;c-KitD814V, and moribund RE;c-KitT417IΔ418-419 mice in M3434 methylcellulose. Each data point represents the total number of myeloid CFU from an independent animal counted 7 to 12 days after plating.
Myeloid-lineage phenotype of leukemic cells in RE;c-KitD814V and RE;c-KitT417IΔ418-419 mice. (A) Representative FACS analysis of AML cells harvested from BM and spleen of Bex;Vex control and moribund RE;c-KitD814V and RE;c-KitT417IΔ418-419 mice. Cells were gated for Bex+Vex+ expression and then analyzed for Mac-1, Gr-1, and CD115 (M-CSFR) expression. (B) Wright-Giemsa–stained cytospins of FACS-purified Bex+Vex+ myeloid-gated BM cells from control or moribund animals demonstrating a high frequency of blast forms in leukemic RE;c-KitD814V and RE;c-KitT417IΔ418-419 mice (original magnification ×630). (C) Myeloid CFU assays performed by plating 1 × 104 FACS-purified Bex+Vex+ myeloid scatter-gated BM cells from Bex;Vex control, nonleukemic RE (39-45 weeks post-transplant), nonleukemic D814V (3-17 weeks post-transplant), moribund RE;c-KitD814V, and moribund RE;c-KitT417IΔ418-419 mice in M3434 methylcellulose. Each data point represents the total number of myeloid CFU from an independent animal counted 7 to 12 days after plating.
The reduced penetrance of the AML phenotype in RE;c-KitT417IΔ418-419 mice compared with RE;c-KitD814V animals that progressed to AML was not because of reduced levels of RE or c-KitT417IΔ418-419 expression compared with levels observed in RE;c-KitD814V mice. Before the onset of changes leading to more aggressive malignancy (preleukemic phase), the mean fluorescence intensity (MFI) of Bex+Vex+ cells in PB of 8 to 10 weeks post-transplant RE;c-KitD814V animals that would ultimately progress to AML was 3151 for Bex (RE expression, n = 8) and 1333 for Vex (c-KitD814V expression, n = 8), whereas the MFI for preleukemic 8 to 10 weeks after transplantation RE;c-KitT417IΔ418-419 mice was 3060 for Bex (RE, n = 18) and 2858 for Vex (c-KitT417IΔ418-419; n = 18; supplemental Figure 2). The difference in RE levels between genotypes was not statistically significant (P = .89, unpaired t test), whereas higher c-Kit levels in RE;c-KitT417IΔ418-419 mice were significant (P = .02, unpaired t test). In addition, the reduced penetrance and delayed onset of AML in RE;c-KitT417IΔ418-419 mice was not because of reduced levels of PB chimerism in preleukemic RE;c-KitT417IΔ418-419 mice compared with age-matched RE;c-KitD814V animals (Bex+Vex+ chimerism levels at 8-10 weeks averaged 5.5% in RE;c-KitT417IΔ418-419 mice that progressed to AML, n = 18, versus 1.6% in RE;c-KitD814V animals, n = 8, P = .04, Mann-Whitney test). These results suggest that intrinsic differences between the distinct subtypes of c-Kit–activating mutations accounted for differences in both penetrance and onset of lethal AML in the 2 models. Because RE and both mutant c-Kit retroviruses would have had an equivalent potential to integrate within loci that could have enhanced AML progression, it is highly unlikely that differences in AML penetrance and kinetics of progression between RE;c-KitD814V and RE;c-KitT417IΔ418-419 mice were the result of cooperating mutations caused by retroviral integration. To address this further, we used both inverse PCR and ligation-mediated PCR to characterize the flanking sequences for a number of retroviral integration events representing AML samples from RE;c-KitD814V or RE;c-KitT417IΔ418-419 mice (Figure 4) and found no common integration sites that might account for differences in AML penetrance or kinetics (supplemental Table 1).
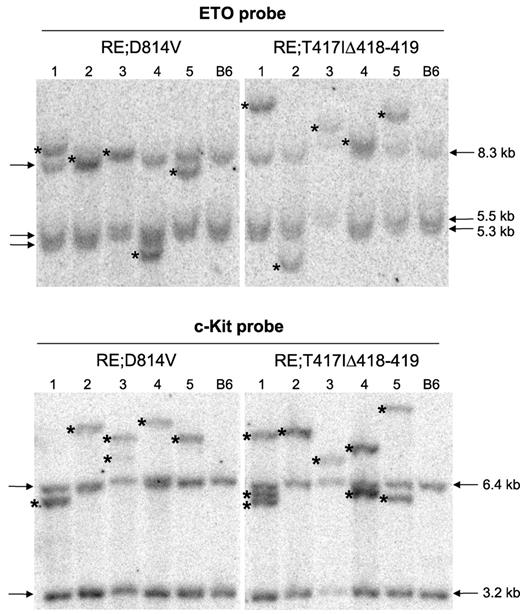
RE;c-KitD814V and RE;c-KitT417IΔ418-419 AML phenotypes are clonal. Southern blot analysis using genomic DNA isolated from whole splenocytes obtained from 5 independent, moribund primary RE;c-KitD814V or RE;c-KitT417IΔ418-419 mice. Lane numbers represent the same DNA sample used for both blots. Blots were hybridized with radiolabeled probes complementary to ETO or c-Kit sequences to detect unique retroviral integrants. Arrows indicate the endogenous murine Eto and c-Kit bands. Wild-type C57BL/6 (B6) splenocytes served as control samples for each blot. Asterisks indicate unique retroviral integrants in each sample.
RE;c-KitD814V and RE;c-KitT417IΔ418-419 AML phenotypes are clonal. Southern blot analysis using genomic DNA isolated from whole splenocytes obtained from 5 independent, moribund primary RE;c-KitD814V or RE;c-KitT417IΔ418-419 mice. Lane numbers represent the same DNA sample used for both blots. Blots were hybridized with radiolabeled probes complementary to ETO or c-Kit sequences to detect unique retroviral integrants. Arrows indicate the endogenous murine Eto and c-Kit bands. Wild-type C57BL/6 (B6) splenocytes served as control samples for each blot. Asterisks indicate unique retroviral integrants in each sample.
Furthermore, the initial (at 8-10 weeks) expression levels of RE or c-KitT417IΔ418-419 between RE;c-KitT417IΔ418-419 mice that progressed to AML and RE;c-KitT417IΔ418-419 mice that never progressed during the one year of observation were not different, indicating that other changes resulted in rapid clonal expansion and progression to AML in a subset of RE;c-KitT417IΔ418-419 mice (MFI at 8-10 weeks for RE;c-KitT417IΔ418-419 mice that progressed to AML was 3060 and 2858 for RE and c-KitT417IΔ418-419, respectively, n = 18; and 2608 and 2003 for RE and c-KitT417IΔ418-419 in age-matched mice that never progressed to AML, n = 15; P = .43 for RE and P = .27 for c-KitT417IΔ418-419, unpaired t test, supplemental Figure 2).
Characteristics of AML observed in RE;c-KitD814V and RE;c-KitT417IΔ418-419 mice
All animals with AML had high blast cell frequencies in BM and showed extensive blast infiltration in lung, liver, and spleen (Figure 2A-B) and splenomegaly (Figure 2C). In addition, lymph node involvement was notable in RE;c-KitT417IΔ418-419 mice (data not shown). We observed a high incidence of granulocytic sarcomas in RE;c-KitD814V animals (7 of 9 mice), which was less common in RE;c-KitT417IΔ418-419 mice (3 of 19 animals). Granulocytic sarcomas all had high frequencies of Mac-1+Gr-1+ and/or Mac-1−/loGr-1+ blasts (supplemental Figure 3). Platelet counts were significantly reduced in moribund animals coexpressing RE with either c-Kit–activating mutation, but in AML cases, only RE;c-KitT417IΔ418-419 mice exhibited significantly reduced RBC counts and hematocrit (Table 2).
Peripheral blood cell counts in reconstituted mice
| Retroviral construct(s); phenotype . | No. of mice analyzed . | WBC, × 109/L . | RBC, × 109/L . | Hematocrit, % . | Platelets, × 109/L . |
|---|---|---|---|---|---|
| Bex;Vex | 13 | 6.39 ± 4.00 | 8.11 ± 2.22 | 34.6 ± 10.0 | 580 ± 295 |
| RE-nonleukemic | 5 | 6.43 ± 4.00 | 8.15 ± 2.12 | 37.2 ± 3.2 | 644 ± 333 |
| D814V-MPN | 3 | > 200* | 4.30 ± 1.28* | 23.4 ± 10.2 | 686 ± 328 |
| D814V-T-ALL | 3 | 3.41 ± 1.72 | 9.90 ± 1.68 | 45.8 ± 11.8 | 360 ± 228 |
| D814V-B-ALL | 1 | 12.34 | 5.07 | 20.9 | 191 |
| T417IΔ418–419-B-ALL | 1 | 80.29 | 6.51 | 29.7 | 270 |
| RE;T417IΔ418–419-AML | 11 | 15.59 ± 19.83 | 2.77 ± 2.12* | 15.6 ± 11.1* | 157 ± 151* |
| RE;D814V-AML | 8 | 11.39 ± 11.55 | 7.11 ± 2.37 | 30.9 ± 9.8 | 142 ± 97* |
| RE;D814V-MPN | 6 | 22.14 ± 15.89* | 3.78 ± 1.47* | 16.9 ± 9.4* | 132 ± 21* |
| RE;D814V-B-ALL | 4 | 19.94 ± 29.64 | 7.20 ± 2.81 | 29.6 ± 8.0 | 140 ± 84* |
| Retroviral construct(s); phenotype . | No. of mice analyzed . | WBC, × 109/L . | RBC, × 109/L . | Hematocrit, % . | Platelets, × 109/L . |
|---|---|---|---|---|---|
| Bex;Vex | 13 | 6.39 ± 4.00 | 8.11 ± 2.22 | 34.6 ± 10.0 | 580 ± 295 |
| RE-nonleukemic | 5 | 6.43 ± 4.00 | 8.15 ± 2.12 | 37.2 ± 3.2 | 644 ± 333 |
| D814V-MPN | 3 | > 200* | 4.30 ± 1.28* | 23.4 ± 10.2 | 686 ± 328 |
| D814V-T-ALL | 3 | 3.41 ± 1.72 | 9.90 ± 1.68 | 45.8 ± 11.8 | 360 ± 228 |
| D814V-B-ALL | 1 | 12.34 | 5.07 | 20.9 | 191 |
| T417IΔ418–419-B-ALL | 1 | 80.29 | 6.51 | 29.7 | 270 |
| RE;T417IΔ418–419-AML | 11 | 15.59 ± 19.83 | 2.77 ± 2.12* | 15.6 ± 11.1* | 157 ± 151* |
| RE;D814V-AML | 8 | 11.39 ± 11.55 | 7.11 ± 2.37 | 30.9 ± 9.8 | 142 ± 97* |
| RE;D814V-MPN | 6 | 22.14 ± 15.89* | 3.78 ± 1.47* | 16.9 ± 9.4* | 132 ± 21* |
| RE;D814V-B-ALL | 4 | 19.94 ± 29.64 | 7.20 ± 2.81 | 29.6 ± 8.0 | 140 ± 84* |
Peripheral blood was collected by cardiac puncture from nonleukemic or moribund mice. A 50-μL sample was subjected to complete blood cell counting using a Hemavet 950 instrument. Data are mean ± SD.
P < .01, compared with Bex;Vex controls (unpaired t test).
FACS analysis of BM and spleen in moribund RE;c-KitD814V and RE;c-KitT417IΔ418-419 mice showed the presence of differing frequencies of Mac-1+Gr-1+ and/or Mac-1−/loGr-1+ blast cells as was noted in the granulocytic sarcomas (supplemental Figure 3, Figure 3A). Interestingly, BM cells expressing RE;c-KitD814V lacked CD115 (M-CSF receptor) expression (Figure 3A), which was a distinguishing marker that characterized AML in RE;c-KitD814V mice from AML in RE;c-KitT417IΔ418-419 animals. CD115+ BM cells in control and RE;c-KitT417IΔ418-419 mice were always Mac-1+Gr-1+ (data not shown). It is not clear whether RE;c-KitD814V coexpression blocked myeloid differentiation before induction of CD115, which seems unlikely because we observed differentiated myeloid cells within BM cytospin preparations from RE;c-KitD814V mice (Figures 2A and 3B), or whether the c-KitD814V mutation blocked expression of the M-CSF receptor at some level. Consistent with reduced CD115 expression, RE;c-KitD814V mice with AML had significantly lower frequencies of monocytes than RE;c-KitT417IΔ418-419 or control mice among Bex+Vex+ cells (Figure 2A). Analysis of 32 RE+ human AML microarray samples showed no significant reduction in CD115 mRNA expression in 9 samples that also possessed c-KITD816V mutations (P. Valk, personal communication, February 2011), which suggests that CD115 expression may be post-transcriptionally regulated in RE;c-KitD814V blasts or our observations are unique to murine cells.
Quantification of myeloid CFU in methylcellulose supplemented with SCF, IL-6, IL-3, and erythropoietin showed that RE alone increased CFU numbers 8- to 10-fold compared with plating equivalent numbers of Bex+Vex+ control BM cells (Figure 3C). Coexpression of RE;c-KitD814V (AML phenotype) or c-KitD814V expression alone completely abrogated myeloid CFU. Curiously, RE;c-KitT417IΔ418-419 expression inhibited myeloid CFU in a fraction (5/8) of RE;c-KitT417IΔ418-419 mice with AML (Figure 3C). In the 3 RE;c-KitT417IΔ418-419 samples where colony numbers were similar to what was observed with RE alone, the average MFI for c-KitT417IΔ418-419 expression was 2738, whereas the average c-Kit MFI for the 5 samples where CFU were blocked was 4342. Although lower c-KitT417IΔ418-419 expression correlated with reduced ability to block myeloid CFU formation, this was not statistically significant (P = .24, unpaired t test).
AML is clonal and transplantable from moribund RE;c-KitD814V and RE;c-KitT417IΔ418-419 mice
The relatively long latency for development of AML in both RE;c-KitD814V and RE;c-KitT417IΔ418-419 mice suggests that other oncogenic changes occur to promote AML progression. To test whether leukemic cells were clonal, Southern blots were generated using genomic DNA isolated from 5 moribund RE;c-KitD814V and RE;c-KitT417IΔ418-419 mice (lanes 1-5, Figure 4). Blots were hybridized with radiolabeled probes complementary to ETO or c-Kit sequences to detect unique retroviral integrants distinct from germline bands observed in control C57BL/6 genomic DNA. As shown in Figure 4, all RE proviral integrants (ETO probe) showed a clonal pattern where one unique integrant (see asterisks) was responsible for all donor hematopoietic cells in the spleen. Analysis of c-Kit mutant retroviral integrants from the same mice showed one or 2 integrants per sample, again indicating clonal expansion of a selected Bex+Vex+ leukemic clone for all moribund RE;c-KitD814V and RE;c-KitT417IΔ418-419 mice.
To test whether AML was transplantable, we transferred varying doses of whole BM cells into lethally irradiated secondary recipient mice. We observed a dose-dependent, rapid (3-10 weeks) progression to lethal AML using equivalent doses of either RE;c-KitD814V– or RE;c-KitT417IΔ418-419–expressing cells (Table 3). FACS profiles of AML cells in hematopoietic tissues of secondary recipient mice were identical to what was observed in the primary recipients (Figure 5). The frequency of leukemia-initiating cells for both RE;c-Kit mutant genotypes was similar, with 100 000 to 500 000 whole BM cells being equivalent to a limiting dilution dose of leukemia-initiating cells (Table 3). It was not possible to analyze leukemia-initiating cell potential in FACS-sorted leukemic HSCs and myeloid progenitor subsets because expression of the murine c-Kit mutations in the context of a retroviral vector could have resulted in c-Kit expression on cells that normally would not express c-Kit, leading to inaccurate phenotyping of cell subsets according to the classic LSK profile.
Transplantability of AML phenotypes into secondary recipient animals
| Cell dosage . | RE;D814V, moribund/total . | RE;T417IΔ418-419, moribund/total . |
|---|---|---|
| 1 × 106 | 4/4 | 6/6 |
| 1.5 × 105 | 8/13 | 8/9 |
| 5 × 104 | 9/14 | 1/5 |
| 1 × 104 | 0/3 | 0/6 |
| Cell dosage . | RE;D814V, moribund/total . | RE;T417IΔ418-419, moribund/total . |
|---|---|---|
| 1 × 106 | 4/4 | 6/6 |
| 1.5 × 105 | 8/13 | 8/9 |
| 5 × 104 | 9/14 | 1/5 |
| 1 × 104 | 0/3 | 0/6 |
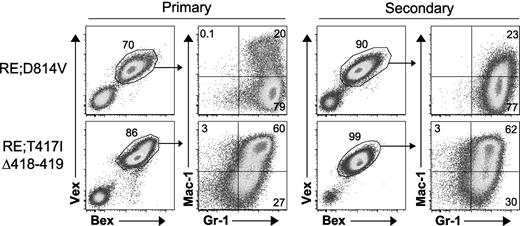
RE;c-KitD814V and RE;c-KitT417IΔ418-419 blasts are malignant in secondary recipients. Whole BM from moribund primary animals (RE;c-KitD814V, n = 4; RE;c-KitT417IΔ418-419, n = 5) was transplanted at various doses (5 × 104 to 1 × 106 cells) into lethally (9 Gy) irradiated secondary hosts. Representative BM flow cytometry profiles of primary and secondary recipient mice. Cells were gated for Bex+Vex+ expression and then analyzed for Mac-1 and Gr-1 expression.
RE;c-KitD814V and RE;c-KitT417IΔ418-419 blasts are malignant in secondary recipients. Whole BM from moribund primary animals (RE;c-KitD814V, n = 4; RE;c-KitT417IΔ418-419, n = 5) was transplanted at various doses (5 × 104 to 1 × 106 cells) into lethally (9 Gy) irradiated secondary hosts. Representative BM flow cytometry profiles of primary and secondary recipient mice. Cells were gated for Bex+Vex+ expression and then analyzed for Mac-1 and Gr-1 expression.
Characterization of MPN and pre-B ALL in RE;c-KitD814V mice
RE;c-KitD814V mice with lethal MPN (35% of mice) had approximately 4-fold higher WBC counts in moribund mice than control animals (22.14 ± 15.89 K/μL versus 6.39 ± 4.0 K/μL, respectively) and showed approximately 2- to 4-fold reductions in hematocrit, RBC, and platelet counts versus controls (Table 2). The increased WBC count was characterized by high percentages of more differentiated Mac-1+Gr-1+ myeloid cells in BM and spleen (Figure 6A; and data not shown) and splenomegaly (supplemental Figure 4A). The splenic architecture was significantly effaced with extensive myeloid infiltrates in the red pulp, as well as infiltrates of more mature myeloid cells in the parenchyma of the lungs and liver (supplemental Figure 4B). The 20% of RE;c-KitD814V mice that presented with pre-B cell leukemias had Bex+Vex+ cells that showed a distinctly lymphocyte forward scatter (FSC) and side scatter (SSC) profile in BM that was readily distinguishable from myeloid scatter profiles observed in all cases of AML or MPN (Figure 6A). The reduced granularity of the lymphoid blasts observed in the FSC/SSC profiles was also noted in cytospin preparations of BM cells from control and moribund RE;c-KitD814V mice with pre-B ALL (Figure 6B). Leukemic pre-B cells expressed B220 and CD19 but did not express IgM on the cell surface (Figure 6A; and data not shown). c-KitD814V animals that developed highly aggressive MPN had even higher WBC counts (> 200 K/μL) than RE;c-KitD814V mice with MPN but normal platelet counts and hematocrit (Table 2), suggesting that RE was primarily responsible for the reduced platelet counts and more severe anemia in RE;c-KitD814V mice. MPN in c-KitD814V animals was Mac-1+Gr-1+CD115− and displayed a myeloid FSC/SSC profile (Figure 6C).
Immunophenotypic and morphologic comparison of neoplastic cells from RE;c-KitD814V mice with AML, MPN, or pre-B-ALL. (A) Representative FACS analysis of neoplastic cells harvested from BM of Bex;Vex control and moribund RE;c-KitD814V mice with the indicated disease phenotype. Cells were gated for Bex+Vex+ expression and then analyzed for FSC/SSC profiles and Mac-1, Gr-1, B220, and CD19 expression. (B) Wright-Giemsa–stained cytospins of FACS-purified Bex+Vex+ myeloid scatter-gated or Bex+Vex+ lymphoid blast scatter-gated BM cells from control or moribund animals (original magnification ×630). Expansion of metamyelocytes, band forms, and eosinophilic progenitors was noted in RE;c-KitD814V mice with MPN. Morphology of pre-B-ALL cells in RE;c-KitD814V mice was predominantly lymphoblastic. (C) FACS analysis of myeloid-lineage surface markers and Wright-Giemsa–stained cytospin preparation of FACS-purified Vex+ myeloid-gated BM cells from a representative moribund animal with c-KitD814V+ MPN showing a predominance of maturing granulocytes.
Immunophenotypic and morphologic comparison of neoplastic cells from RE;c-KitD814V mice with AML, MPN, or pre-B-ALL. (A) Representative FACS analysis of neoplastic cells harvested from BM of Bex;Vex control and moribund RE;c-KitD814V mice with the indicated disease phenotype. Cells were gated for Bex+Vex+ expression and then analyzed for FSC/SSC profiles and Mac-1, Gr-1, B220, and CD19 expression. (B) Wright-Giemsa–stained cytospins of FACS-purified Bex+Vex+ myeloid scatter-gated or Bex+Vex+ lymphoid blast scatter-gated BM cells from control or moribund animals (original magnification ×630). Expansion of metamyelocytes, band forms, and eosinophilic progenitors was noted in RE;c-KitD814V mice with MPN. Morphology of pre-B-ALL cells in RE;c-KitD814V mice was predominantly lymphoblastic. (C) FACS analysis of myeloid-lineage surface markers and Wright-Giemsa–stained cytospin preparation of FACS-purified Vex+ myeloid-gated BM cells from a representative moribund animal with c-KitD814V+ MPN showing a predominance of maturing granulocytes.
Discussion
In this study, we compared the ability of 2 classes of c-Kit–activating mutations to cooperate with RUNX1-ETO in promotion of AML. RE;c-KitD814V or RE;c-KitT417IΔ418-419 mice both developed AML, although with different kinetics and penetrance (Figure 1C). Consistent with clinical observations showing higher coincident expression of c-KITD816V with RUNX1-ETO (11%-44% of cases) than exon 8 mutations, such as c-KITT417IΔ418-419 (2%-13% of cases), we observed more robust progression to AML in RE;c-KitD814V mice than RE;c-KitT417IΔ418-419 animals (Figure 1C-D).24-30,32,33,38 One explanation for the delayed onset and incomplete penetrance of AML observed in RE;c-KitT417IΔ418-419 mice could be reduced potency and/or altered signaling associated with mutations in the extracellular domain of c-Kit. Previous studies have shown that c-Kit exon 8 mutations promote spontaneous receptor dimerization, hyper-responsiveness to SCF, and growth factor-independent proliferation of IL-3–dependent cell lines, such as Ba/F3 and FDC-P1.38,45 In one study,38 3 representative c-Kit exon 8 mutant alleles were not autophosphorylated, although significant autophosphorylation of a similar exon 8 receptor mutant was observed by another group.46 The latter group also noted that differing classes of c-Kit mutation (extracellular vs activation loop) activate both overlapping and distinct signaling pathways downstream of c-Kit.46 This may be responsible for the overlap, and yet distinct difference, between gene expression signatures in human AML blasts with the CBF mutations [t(8;21)(q22;q22) or inv(16)(p13.1;q22)] and c-Kit–activating mutations in either exon 8 or exon 17.47
In addition to inherent differences in the signaling potential of distinct classes of c-Kit–activating mutations, it is also possible that alteration in c-Kit localization between RE;c-KitT417IΔ418-419 and RE;c-KitD814V mice might have contributed to the observed variability in leukemic penetrance. As noted in Figure 1B, the level of intracellular c-KitD814V protein was only approximately 3-fold lower than the level of c-KitT417IΔ418-419 protein. However, cell-surface expression of c-KitD814V was approximately 50-fold lower than c-KitT417IΔ418-419 protein. This difference in localization between the 2 c-Kit mutations could be explained by altered intracellular trafficking that was specific for the c-KitD814V mutation40,48 or enhanced degradation of c-KitD814V in the absence of SCF.49 The impact of this difference on phenotype is not clear because c-KitD814V would still have had transforming activity when localized to the Golgi, as shown in studies using a domain-tagged mouse-human hybrid c-KITD816V allele that induced MPN in mice.40 Because we did not observe a significant correlation between the levels of mutant c-Kit expression in AML blasts (in vivo MFI determinations) and the ability of each c-Kit mutant to stimulate more rapid progression to AML (the highest expression was seen for the less potent c-KitT417IΔ418-419 allele), it seems unlikely that differences in expression level between the c-Kit mutants would account for the phenotypic differences in the animals.
The relatively long latency (Figure 1C; supplemental Figure 1) and the clonal nature of the leukemic blasts in RE;c-KitD814V and RE;c-KitT417IΔ418-419 mice that progressed to AML (Figure 4) suggest that additional oncogenic changes were occurring that were essential for leukemic progression in the context of RE and both classes of c-Kit–activating mutations. This is likely true for other receptor tyrosine kinase mutations, such as FLT3-ITD, which weakly cooperates with RE to promote leukemia with a mean latency of 233 days since retroviral integration patterns were monoclonal/oligoclonal in this model.20 Mice that developed AML within 1 to 4.5 months when RE was coexpressed with Wilms tumor protein (WT1) also exhibited monoclonal/oligoclonal hematopoiesis, whereas hematopoiesis was polyclonal among cells that only expressed RE.21 A possible exception to the need for additional oncogenic events for AML progression may be RE coexpression with a TEL/PDGRβR fusion, where a rapidly lethal AML was observed within 2 months of reconstitution in 19 of 19 mice and blasts were oligoclonal based on retroviral integration patterns.19 Although 60% to 70% of human t(8;21)+ samples have nonoverlapping RTK or activating NRAS or KRAS mutations, additional recurrent genetic abnormalities have also been noted in a high percentage of cases, including loss of a sex chromosome in approximately 50% of cases and deletions in 9q,31 which indicates that additional oncogenic changes that may cooperate with RE during AML progression remain to be identified.
An interesting difference between the mouse modeling of RE;c-KitD814V and human patients with RE and c-KITD816V lesions is the broad spectrum of both lymphoid and myeloid malignancies observed in transplanted mice compared with the exclusively AML phenotype seen in humans. One probable explanation for this difference relates to the temporal order in which mutations occur during leukemic progression in humans, which is not typically modeled in murine studies using retroviral or gene-targeted approaches. Sequential occurrence of oncogenic insults may limit the target cell that secondary AML-promoting events occur in if the primary event restricts developmental potential, as is the case with RUNX1-ETO, which efficiently blocks early T-cell development resulting in the absence of RUNX1-ETO transcripts in T cells in RUNX1-ETO knock-in mice or in human t(8;21)+ patient samples.14,50 The exclusive AML phenotype in humans may also be the result of the dramatic expansion in myeloid progenitor cells in BM driven by RE in the preleukemic phase,16 which would increase the likelihood for subsequent AML-promoting changes (eg, activating RTK mutations) to randomly occur in myeloid progenitor cells compared with lymphoid progenitors, which do not expand with RE. This expansion in myeloid progenitor cells is not modeled when 2 mutations are simultaneously introduced into HSC/progenitors cells used to reconstitute mice. There is reasonably good evidence that the RUNX1-ETO translocation is a primary event in leukemogenesis based on observations that RE can be detected in utero at relatively high frequencies13 and observations showing that patients diagnosed with both RE and c-KIT activating mutations only retain RE+ cells during clinical remission.30 This could lead to myeloid progenitor cell expansion in human BM during a t(8;21)+ preleukemic phase that could predispose to further progression-associated changes within the myeloid lineage.
Another simple explanation for differences between the murine and human phenotypes related specifically to expression of the c-KitD814V mutation with RE is that the retrovirus is expressing c-KitD814V in lymphoid cells when the endogenous c-Kit locus would normally be inactive (c-Kit expression is down-regulated at very early stages in both B- and T-cell development, and this would not be the case using retroviral vectors). However, this does not explain why the c-KitT417IΔ418-419 mutant, even though it is expressed at higher total protein levels than c-KitD814V (Figure 1B), does not show any lymphoid or MPN phenotypes when coexpressed with RE. It is possible that inherent signaling differences between the 2 mutant c-Kit receptors were driving this outcome, although this remains to be formally investigated. What was clear is that RE expression dramatically altered the leukemic potential of both c-KitD814V and c-KitT417IΔ418-419, which resulted exclusively in lymphoid leukemias or MPN when expressed alone, toward AML when they were coexpressed with RE (Table 1).
The results of this study functionally demonstrate that 2 major classes of c-Kit–activating mutations can cooperate with RE to promote AML in mice. In a very recent study, 2 different c-Kit mutants were coexpressed with RE in a retroviral transduction/transplantation model.37 One c-Kit mutant in this study was representative of a juxtamembrane c-Kit mutation encoded by exon 11, whereas the other (N822K) is a commonly observed activation loop mutant that is distinct from c-KitD814V in that it retains sensitivity to imatinib.37 Similar to what we observed in c-KitD814V mice, animals expressing only c-KitN822K developed lethal myeloproliferative disease. When coexpressed with RE, RE;c-KitN822K mice developed a transplantable AML with a median latency of 177 days in approximately 70% of reconstituted mice, which contrasts with the 100% penetrance that we observed at a median latency of 94 days for RE;c-KitD814V mice that developed AML (Figure 1D). Curiously, the RE;c-KitN822K mice in their study more closely resembled the malignant phenotype of RE;c-KitT417IΔ418-419 animals with respect to leukemic penetrance and uniquely AML presentation. Together, these analyses demonstrate that the 3 most common activating c-KIT mutations observed in RE+ patient samples function as driver mutations that promote progression to AML. Further work will be necessary to define the entire spectrum of changes that promote progression to AML in conjunction with RE.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Larry Gartland for expert cell sorting; Dr Peter Valk, Erasmus University, Rotterdam, The Netherlands, for information regarding microarray studies of human AML samples; and members of the C.A.K. laboratory for many helpful discussions.
This work was supported by the National Institutes of Health/National Cancer Institute (RO1CA096798 and RO1CA144248).
National Institutes of Health
Authorship
Contribution: H.J.N. and C.A.K. designed the research and wrote the manuscript; H.J.N., H.-G.K., C.-W.C., and V.R. performed the research; and K.W.H. provided critical reagents.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christopher A. Klug, Department of Microbiology, University of Alabama-Birmingham, 1825 University Blvd, Shelby Rm 510, Birmingham, AL 35294-2182; e-mail: chrisk@uab.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal