Abstract
In vivo lentiviral vector (LV)–mediated gene delivery would represent a great step forward in the field of gene therapy. Therefore, we have engineered a novel LV displaying SCF and a mutant cat endogenous retroviral glycoprotein, RDTR. These RDTR/SCF-LVs outperformed RDTR-LVs for transduction of human CD34+ cells (hCD34+). For in vivo gene therapy, these novel RDTR/SCF-displaying LVs can distinguish between the target hCD34+ cells of interest and nontarget cells. Indeed, they selectively targeted transduction to 30%-40% of the hCD34+ cells in cord blood mononuclear cells and in the unfractionated BM of healthy and Fanconi anemia donors, resulting in the correction of CD34+ cells in the patients. Moreover, RDTR/SCF-LVs targeted transduction to CD34+ cells with 95-fold selectivity compared with T cells in total cord blood. Remarkably, in vivo injection of the RDTR/SCF-LVs into the BM cavity of humanized mice resulted in the highly selective transduction of candidate hCD34+Lin− HSCs. In conclusion, this new LV will facilitate HSC-based gene therapy by directly targeting these primitive cells in BM aspirates or total cord blood. Most importantly, in the future, RDTR/SCF-LVs might completely obviate ex vivo handling and simplify gene therapy for many hematopoietic defects because of their applicability to direct in vivo inoculation.
Introduction
Gene therapy has proven to be promising as a cure for many inherited and acquired diseases, as evidenced by the successful treatment of immunodeficiencies.1,2 Recently, Cartier et al for the first time used lentiviral vectors (LVs) in a successful clinical trial on the treatment of adrenoleukodystrophy.3 For the correction of these defects, the therapeutic gene must be delivered to cells able to both self-renew and differentiate into any hematopoietic lineage. Therefore, these gene therapies must be targeted to the “right” cell, the human hematopoietic stem cell (hHSC), without modifying its properties. Ex vivo–targeted HSC gene delivery is associated with a risk of inducing cell differentiation and loss of the homing/engraftment potential of these cells.4,5 In contrast, in vivo gene transfer could target HSCs in their stem cell niche, a microenvironment that regulates HSC survival and maintenance.6 A minimal number of gene-corrected cells needs to be reinfused, and this requirement could be a limiting factor in BM failure syndromes such as Fanconi anemia (FA), in which patients already suffer from a reduced number of human CD34+ (hCD34+) cells.7-9 This important limitation for FA ex vivo gene therapy might be overcome by in vivo targeted gene delivery to HSCs.10 A major barrier in LV transduction of HSCs is that the majority of HSCs are in the G0 phase of the cell cycle and only occasionally undergo self-renewing cell divisions11 ; HSCs in the G0 phase are also poorly permissive to classic VSV-G–LV transduction. Clearly, short cytokine stimulation induces high transduction levels in HSCs.12,13 We previously engineered “early-activating cytokine”–displaying LVs that allowed a slight and transient stimulation of hCD34+ cells, resulting in efficient lentiviral gene transfer while preserving the “stemness” of the targeted HSCs.13 Indeed, thrombopoietin (TPO) and SCF, two cytokines that allow cells to retain comparable engraftment abilities with respect to unmanipulated HSCs, were displayed at the LV surface.14,15 The selective transduction of HSCs by these VSV-G/TPO/SCF–displaying vectors was demonstrated by their capacity to promote selective transduction of long-term NOD/SCID repopulating cells.13
Two major challenges for in vivo LV gene delivery are exposure to the host immune/complement system and off-target cell gene transfer after systemic administration. To surmount these obstacles, complement-resistant vectors capable of specifically targeting the very rare immature progenitor cells are required. Indeed, because the fusion glycoprotein VSV-G in VSV-G/TPO/SCF–co-displaying LVs is sensitive to human complement and its receptor is present on all tissues, these LVs are unsuited for in vivo gene delivery.16,17 Therefore, we present here a second generation of early-acting-cytokine–displaying LVs fit for targeted in vivo gene delivery to hCD34+ cells. To achieve this, the VSVG glycoprotein was exchanged for another fusion partner, a mutant RD114 glycoprotein of an endogenous feline virus, RD114 (Figure 1A). RD114 is an attractive candidate for in vivo use because: (1) RD114 glycoprotein is resistant to degradation by human complement17 ; (2) RD114-pseudotyped LVs efficiently transduce CD34+HSCs17-19 ; and (3) a RD114 glycoprotein mutant (RDTR) allowing incorporation onto LVs has been engineered.17
We report here that RDTR/SCFHA–displaying vectors achieve a highly selective transduction of hCD34+ cells in unfractionated total cord blood (CB) and BM. Remarkably, these novel LVs targeted gene transfer to hCD34+ cells in vivo in the BM of humanized Balbc rag2−/−γc−/− mice. Moreover, this new generation of LVs targeted gene transfer to the reduced portion of hHSCs in unfractionated BM from FA patients. Therefore, this vector provides improved alternatives for gene therapy of BM failure syndromes both ex vivo and, in the future, in vivo.
Methods
Envelope construction
The TPO171HA chimeric envelope glycoprotein (here called TPOHA) has been described previously.13 The SfiI/NotI fragment from SCFSU13 was fused to the influenza hemagglutinin (HA) env gene using the SfiI/NotI backbone fragment of TPOHA, resulting in the SCFHA glycoprotein. RDTR is a mutated feline retrovirus (RD114) glycoprotein that has been described previously.17 All chimeric env glycoproteins were expressed in the phCMV-G expression vector backbone.20
Production of retroviral vectors and titration
Self-inactivating HIV-1–derived vectors were generated as described previously by transfection of 293T cells.20 Briefly, for HIV vector production, cotransfection was performed with the Gag-Pol packaging construct 8.91 and the self-inactivating (SIN) HIV-1–derived vector coding for the green fluorescent protein (GFP) reporter under the control of a SFFV promoter (SIN-HIVSFFVGFP), as described previously.21 For co-display of RDTR with SCFHA and/or TPOHA, 7 μg of RDTR and 2 μg of the cytokine-fused glycoproteins were transfected. Together with the chimeric env glycoproteins, a plasmid encoding neuraminidase was transfected.17 At 16 hours after transduction, the DMEM was replaced by serum-free CellGro medium (Cellgenix) and 36 hours after transfection, vectors were harvested, filtrated, and concentrated by filtration (Sartorius). Titration was performed by adding serial dilutions of vector to 293T cells and the multiplicities of infection (MOIs) were expressed using 293T titers.
Determination of activity of cytokine-displaying vectors
The functional activity of SCFHA-displaying vectors was evaluated on BAF3-cKit cells, and the BAF3-Mpl cells were used to evaluate the activity of TPOHA-displaying vectors, as described previously13 and in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Western blot analysis and immunoprecipitation assay
Western blot analysis was performed as described previously.22 The immunoprecipitation assay was performed by incubating 1 mL of fresh, unconcentrated vector supernatant with anti-RD114 Ab or anti-HA Ab (rabbit antiserum against H7 was provided by W. Garten, Marburg, Germany) at 2μg/mL according to the manufacturer's instructions (Ademtech), followed by Western blotting of precipitated viral particles as described previously.20
Sample collection and isolation of CD34+ cells
Sample collection and isolation of CD34+ cells were performed as described previously.13
Transduction assays
CD34+ CB cells (5 × 104) were transduced in 48-well plates with fresh or concentrated LVs at MOIs of 1, 10, or 100 (as indicated) in CellGro serum-free medium. Where indicated, transductions were performed on RetroNectin (TAKARA)–coated plates according to the manufacturer's instructions. Transduction of CB or BM mononuclear cells (MCs) was performed by transduction of 1 × 105 total MCs/well in CellGro medium seeded in a 48-well plate at the indicated vector doses and in the absence or presence of RetroNectin. The protocol to evaluate stable and selective hCD34+-cell transduction in total CB MCs is outlined in Figure 2C. The transduction protocol of whole CB is outlined in Figure 4A. For transduction of unselected BM cells from FA patients, cells were resuspended in CellGro supplemented with 5 μg/mL of Eternacept (Immunex) and 5 ng/mL of recombinant human TPO.
Conditioning and reconstitution of Balbc rag2−/−γc−/− mice
Balbc rag2−/−γc−/− immunodeficient mice were obtained from Dr Mamoro Ito and Taconic (Central Institute for Experimental Animals, Kawasaki, Japan). Experiments were performed in accordance with the guidelines of institutional animal care after approval of protocols by the ethical committee. The 2- to 4-day-old newborn BALB/c Rag2−/−γc−/− mice were subjected to a sublethal irradiation of 2 × 1.5 Gy. For evaluation of reconstitution/differentiation capacity of RDTR/SCFHA-LV–transduced human CB (hCB) CD34+ cells, 2 × 105 cells were injected intrahepatically into newborn mice. To evaluate the in vivo performance of RDTR/SCFHA LVs, 2 × 105 freshly isolated, untransduced cells were injected intrahepatically into newborn mice. From week 13 on, blood was taken from the facial vein to verify the reconstitution (ie, the percentage of human CD45+ [hCD45+] cells) by FACS.
Intrafemoral vector injection of Balbc Rag2−/−γc−/− mice
The mice were anesthetized by IP injection of a mixture of 70:30 ketamine:xylasine. A small incision was made at the top of the kneecap using a pair of small scissors. The femur was pierced with a needle, the animals were injected with a maximum of 10 μL of vector (titers = 3-10 × 107 transducing units/mL) in the femoral lower epiphysis, and the incision was stitched up. Mice were monitored during the waking-up period and for 1 week after the injection. An analgesic drug was added to the drinking water after surgery.
FACS analysis
See supplemental Methods for a description of the FACS analysis.
DNA extraction and quantitative PCR
DNA extraction and quantitative PCR were performed as described previously.23
Results
RDTR/SCFHA-displaying LVs promote high-level gene transfer in immature CB cKit+hCD34+ cells
Previously, we developed VSV-G/TPO- and VSV-G/SCF-LVs that outperformed conventional VSV-G–LVs for ex vivo gene delivery to the most immature HSCs.13 However, because the fusion glycoprotein VSV-G is unsuitable for in vivo targeting, we exchanged it for another fusion partner, the mutant cat endogenous viral RD114 glycoprotein, called hereafter RDTR (Figure 1A).17
Co-display of RDTR and SCF on LVs allows transduction of immature c-Kit+hCD34+ cells. (A) Schematic representation of the LVs displaying the RD114 glycoprotein, SCF, and TPO. A TPO truncated form, 171 aa long, was fused to the N-terminus of the influenza HA glycoprotein (TPOHA). SCF was also fused to the N-terminus of the HA glycoprotein, which allowed efficient functional incorporation on LVs (SCFHA). Because these chimeric HA glycoproteins demonstrated a reduced infectivity, we needed to coexpress an additional fusion competent glycoprotein, RD114. The cytoplasmic tail of RD114 was exchanged for that of the MLV glycoprotein, resulting in a mutant RDTR, which allowed efficient incorporation on HIV vectors as described previously.17 (B) Immunoblots of LV particles displaying RDTR together with TPOHA, SCFHA, or both chimeric glycoproteins at their surface. LVs were purified over a sucrose cushion by ultracentrifugation. The upper part of the membrane was stained with Abs against the surface domain of RD114, the middle part with Abs against the influenza HA glycoprotein to detect the TPOHA and SCFHA chimeric envelopes. HIV-1 capsid (CA) was detected to assess equivalent loading of vectors. (C) CD34+ CB cells were incubated with RDTR/TPO-displaying (RDTR/TPOHA), RDTR/SCF-displaying (RDTR/SCFHA), or RDTR/SCF/TPO-displaying (RDTR/TPOHA/SCFHA) LVs in the presence or absence of RetroNectin. As controls, CD34+ cells were incubated with LVs displaying RDTR in the presence of recombinant cytokines with or without RetroNectin at MOI = 100. At day 3 after transduction, cells were evaluated for CD34+ surface marker and GFP expression by FACS (means ± SD, n = 6). (D) CD34+ CB cells were incubated with HIV vectors co-displaying the RDTR envelope with TPOHA, SCFHA, or both cytokines displaying glycoproteins in the absence of RetroNectin. Counterpart transductions with VSV-G/TPOHA, VSV-G/SCFHA, or VSV-G/SCFHA/TPOHA were performed. Transductions were performed at a MOI = 100 or MOI = 1. The percentage of GFP+CD34+ cells was analyzed by FACS (means ± SD, n = 4). CB hCD34+ cells were pre-incubated with increasing concentrations of rSCF (E) or empty SCFHA displaying lentiviral particles (F) as indicated on the x-axis. We indicated in the x-axis the functional SCF equivalent (= ng SCF activity; see “Determination of activity of cytokine-displaying vectors”). Subsequently, incubation with RDTR/SCFHA-LVs at MOI = 10 was performed. The percentage of GFP+CD34+ cells was analyzed 3 days after transduction by FACS (means ± SD, n = 3). (G) CD34+ CB cells were incubated with RDTR/SCFHA co-displaying vectors; RDTR-LVs in the presence of rSCF, SCFHA, or SCFHAX single-displaying vectors; or a mixture of RDTR single-displaying LVs (RDTR) and SCFHA single-displaying vectors (SCFHA). SCFHAX contains an inactivated cleavage site, making the glycoprotein incompetent for cell fusion. All transductions were performed at MOI = 10 in the presence and absence of RetroNectin. At day 5 after transduction, cells were evaluated for CD34+ surface marker and GFP expression by FACS (means ± SD, n = 3). (H) Immunoprecipitation of RDTR/SCFHA-LVs with anti-HA Abs followed by immunoblot detection of RDTR (left) and immunoprecipitation of RDTR/SCFHA LVs with anti-RD114 Abs followed by detection of SCFHA by Western blot (right) are shown. The positions of RDTR and SCFHA are indicated.
Co-display of RDTR and SCF on LVs allows transduction of immature c-Kit+hCD34+ cells. (A) Schematic representation of the LVs displaying the RD114 glycoprotein, SCF, and TPO. A TPO truncated form, 171 aa long, was fused to the N-terminus of the influenza HA glycoprotein (TPOHA). SCF was also fused to the N-terminus of the HA glycoprotein, which allowed efficient functional incorporation on LVs (SCFHA). Because these chimeric HA glycoproteins demonstrated a reduced infectivity, we needed to coexpress an additional fusion competent glycoprotein, RD114. The cytoplasmic tail of RD114 was exchanged for that of the MLV glycoprotein, resulting in a mutant RDTR, which allowed efficient incorporation on HIV vectors as described previously.17 (B) Immunoblots of LV particles displaying RDTR together with TPOHA, SCFHA, or both chimeric glycoproteins at their surface. LVs were purified over a sucrose cushion by ultracentrifugation. The upper part of the membrane was stained with Abs against the surface domain of RD114, the middle part with Abs against the influenza HA glycoprotein to detect the TPOHA and SCFHA chimeric envelopes. HIV-1 capsid (CA) was detected to assess equivalent loading of vectors. (C) CD34+ CB cells were incubated with RDTR/TPO-displaying (RDTR/TPOHA), RDTR/SCF-displaying (RDTR/SCFHA), or RDTR/SCF/TPO-displaying (RDTR/TPOHA/SCFHA) LVs in the presence or absence of RetroNectin. As controls, CD34+ cells were incubated with LVs displaying RDTR in the presence of recombinant cytokines with or without RetroNectin at MOI = 100. At day 3 after transduction, cells were evaluated for CD34+ surface marker and GFP expression by FACS (means ± SD, n = 6). (D) CD34+ CB cells were incubated with HIV vectors co-displaying the RDTR envelope with TPOHA, SCFHA, or both cytokines displaying glycoproteins in the absence of RetroNectin. Counterpart transductions with VSV-G/TPOHA, VSV-G/SCFHA, or VSV-G/SCFHA/TPOHA were performed. Transductions were performed at a MOI = 100 or MOI = 1. The percentage of GFP+CD34+ cells was analyzed by FACS (means ± SD, n = 4). CB hCD34+ cells were pre-incubated with increasing concentrations of rSCF (E) or empty SCFHA displaying lentiviral particles (F) as indicated on the x-axis. We indicated in the x-axis the functional SCF equivalent (= ng SCF activity; see “Determination of activity of cytokine-displaying vectors”). Subsequently, incubation with RDTR/SCFHA-LVs at MOI = 10 was performed. The percentage of GFP+CD34+ cells was analyzed 3 days after transduction by FACS (means ± SD, n = 3). (G) CD34+ CB cells were incubated with RDTR/SCFHA co-displaying vectors; RDTR-LVs in the presence of rSCF, SCFHA, or SCFHAX single-displaying vectors; or a mixture of RDTR single-displaying LVs (RDTR) and SCFHA single-displaying vectors (SCFHA). SCFHAX contains an inactivated cleavage site, making the glycoprotein incompetent for cell fusion. All transductions were performed at MOI = 10 in the presence and absence of RetroNectin. At day 5 after transduction, cells were evaluated for CD34+ surface marker and GFP expression by FACS (means ± SD, n = 3). (H) Immunoprecipitation of RDTR/SCFHA-LVs with anti-HA Abs followed by immunoblot detection of RDTR (left) and immunoprecipitation of RDTR/SCFHA LVs with anti-RD114 Abs followed by detection of SCFHA by Western blot (right) are shown. The positions of RDTR and SCFHA are indicated.
TPO and SCF were fused to the N-terminus of the influenza HA glycoprotein because this resulted in an efficient co-incorporation with RDTR on LVs (Figure 1A-B). We obtained concentrated titers of 1 × 107 to 1 × 108 IU/mL that were comparable to RDTR-LVs. Functional co-display of TPO and SCF with RDTR on LVs was confirmed on Mpl- and c-Kit–expressing cell lines (data not shown). First, we tested these early-acting cytokine-displaying vectors for the transduction of hCD34+ cells from CB. In the absence of RetroNectin, the resulting RDTR/SCFHA- and RDTR/SCFHA/TPOHA LVs were far more efficient in transducing CB-derived hCD34+ cells than the RDTR-LVs, regardless of the presence of the counterpart cytokines in their soluble form (Figure 1C). This shows that the RDTR-LVs are completely dependent on RetroNectin for the transduction of hCD34+ cells. Importantly for in vivo applications, RDTR/SCFHA-LVs, and to a lesser extent RDTR/TPOHA- or RDTR/SCFHA/TPOHA-LVs, were independent of RetroNectin for transduction of CD34+ cells (Figure 1C). Another important issue for in vivo use is that systemic administration of a vector will result in an important dilution of vector concentration. In the absence of RetroNectin, the RDTR/SCFHA LVs allowed the reduction of vector doses to MOI = 1, without a significant decrease in CD34+-cell transduction efficiency (from 30% to 20%; Figure 1D). RDTR/SCFHA-LVs gave the highest transduction levels for hCD34+ cells compared with the other RDTR/cytokine–displaying LVs. Indeed, the RDTR/TPOHA- and RDTR/TPOHA/SCFHA-LVs resulted in a more than 3-fold reduction of the CD34+ transduction level when MOI = 1 was used compared with MOI = 100. It should be noted that the cytokine-displaying VSV-G/TPOHA-, VSV-G/TPOHA/SCFHA-, and VSV-G/SCFHA-LVs were less suitable for in vivo use because low vector doses resulted in inefficient transduction (Figure 1D).
Our objective was to transduce the most immature subset in the hCD34+ cell population. To demonstrate that RDTR/SCFHA-LV transduction of hCD34+ cells was mediated by the binding of SCF displayed on the vector to its receptor, c-Kit, a receptor expressed on HSCs,24 we performed a competition assay. Preincubation of hCD34+ cells with recombinant SCF (rSCF; Figure 1E) or “empty” LV particles displaying only SCF at their surface (Figure 1F), followed by transduction with RDTR/SCFHA-LVs, resulted in an up to 4-fold reduction of transduction efficiency.
Additional data indicated that co-displaying RDTR and SCF on the same LV particle was absolutely required to obtain efficient transduction of hCD34+ cells for the following reasons: (1) transduction with RDTR/SCFHA-LVs was independent of RetroNectin, whereas this was not the case for RDTR-LVs in presence of rSCF (Figure 1C,G); (2) a mixture of RDTR-LVs and SCFHA-LVs did not allow efficient transduction (Figure 1G); and (3) SCFHA LVs did not allow transduction of CD34+ cells (Figure 1G). Even in the presence of fibronectin, RDTR/SCFHA-LVs demonstrated a much higher transduction efficiency of hCD34+ cells compared with combining RDTR-LVs with SCFHA-LVs in trans. In fact, the transduction behavior of the latter is equivalent to RDTR-LVs in the presence of rSCF (Figure 1G). This enforces our hypothesis that both RDTR and SCFHA need to be incorporated on the same lentiviral particle to achieve selective hCD34+-cell transduction. Therefore, we checked the co-incorporation of both glycoproteins on the same LV by immunoprecipitation of the RDTR/SCFHA–displaying LVs with an anti-RD114 Ab and demonstrated that the SCFHA glycoprotein was present in this fraction (Figure 1H). This result was confirmed by immunoprecipitation of the vectors with anti-HA Ab and detection of RDTR glycoprotein in this fraction (Figure 1H).
These data suggest that the SCFHA and RDTR glycoproteins are co-incorporated into the same vector particle, allowing them to bind to c-Kit and efficiently transduce very immature c-Kit+hCD34+ cells. Therefore, the RDTR/SCFHA-LV was the candidate of choice for further evaluation of targeted gene transfer in HSCs.
RDTR/SCFHA-LVs preferentially target transduction to hCD34+ cells in a heterogeneous population of CB MCs
LVs for in vivo gene therapy use need to be able to distinguish between the nontarget cells and the target cells of interest for gene transfer. The selectivity of RDTR/SCFHA-LVs for hCD34+-cell transduction was evaluated by incubating the vector with CB MCs containing, at the most, 1%-2% hCD34+ cells. The RDTR/SCFHA-LVs were able to transduce preferentially the hCD34+ target cells even at low vector doses (MOI = 1), in contrast to the RDTR LVs + rSCF, which resulted in very poor transduction (Figure 2A). Moreover, the nontarget T cells (CD3+; 80% of the MC population) were very poorly transduced by the RDTR/SCFHA-LVs (Figure 2B-C). Other cell lineages present in the MC population, such as monocytes, B cells, and NK cells, remained untransduced (Figure 2B-C). RDTR/SCFHA-LV transduction of the CD34+ cells using MOI = 1 or MOI = 10 resulted in slightly different transduction levels, but both vector doses resulted in a high SFFV-driven GFP expression intensity (mean fluorescence intensity in Figure 2B and data not shown), most likely because the integrated copy number using both vector doses did not differ significantly and because we used a strong internal promoter in the LV, the SFFV promoter.
RDTR/SCFHA-LVs target gene transfer to hCD34+ cells in the total CB MC population. MCs were isolated from fresh CB using a Ficoll gradient and cultured in the absence of RetroNectin. MCs were incubated with LVs pseudotyped with RDTR or VSV-G in the presence of human rSCF (50 ng/mL) or with vectors co-displaying RDTR or VSV-G with the SCFHA glycoprotein without addition of cytokines at MOI = 10 (A,C) and/or MOI = 1 (A-B). (A) At day 3 after transduction, cells were evaluated for CD34+ surface marker and GFP expression by FACS (means ± SD, n = 4). (B) The percentage of GFP+ cells in early progenitors (CD34+ cells), T cells (CD3+ cells), B cells (CD19+ cells), and natural killer cells (CD56+ cells) in the transduced MC population is indicated. MOI = 1 was used and the mean fluorescence intensities (MFIs) are indicated. The data are representative of 3 experiments. (C) Comparison of the transduction efficiencies between the different lineages in the CB MC population transduced by RD/SCFHA LVs or VSV-G LVs. After 48 hours of transduction, early progenitor cells (CD34+ cells), T cells (CD3+ cells), B cells (CD19+ cells), and monocytes (CD14+ cells) were isolated from the MC fraction, each cell lineage was continued in culture for 6 days as indicated, and transduction was analyzed by FACS (means ± SD, n = 3).
RDTR/SCFHA-LVs target gene transfer to hCD34+ cells in the total CB MC population. MCs were isolated from fresh CB using a Ficoll gradient and cultured in the absence of RetroNectin. MCs were incubated with LVs pseudotyped with RDTR or VSV-G in the presence of human rSCF (50 ng/mL) or with vectors co-displaying RDTR or VSV-G with the SCFHA glycoprotein without addition of cytokines at MOI = 10 (A,C) and/or MOI = 1 (A-B). (A) At day 3 after transduction, cells were evaluated for CD34+ surface marker and GFP expression by FACS (means ± SD, n = 4). (B) The percentage of GFP+ cells in early progenitors (CD34+ cells), T cells (CD3+ cells), B cells (CD19+ cells), and natural killer cells (CD56+ cells) in the transduced MC population is indicated. MOI = 1 was used and the mean fluorescence intensities (MFIs) are indicated. The data are representative of 3 experiments. (C) Comparison of the transduction efficiencies between the different lineages in the CB MC population transduced by RD/SCFHA LVs or VSV-G LVs. After 48 hours of transduction, early progenitor cells (CD34+ cells), T cells (CD3+ cells), B cells (CD19+ cells), and monocytes (CD14+ cells) were isolated from the MC fraction, each cell lineage was continued in culture for 6 days as indicated, and transduction was analyzed by FACS (means ± SD, n = 3).
After a 48-hour transduction of the total CB MC population with the different vectors, we separated CD34+ cells, T cells, B cells, and monocytes by positive selection and cultured them for 6 days, as outlined in Figure 2C. These results clearly underline the selective transduction of hCD34+ cells with RDTR/SCF LVs, in contrast to VSV-G/SCF-LVs, which also allows the transduction of hCD34+ cells, T cells, and monocytes. The VSV-G-LV transduction of T cells was low because TCR stimulation was applied after transduction. Indeed, T cells normally require prestimulation through the TCR to obtain high-level transduction.25
Interestingly, the RDTR/SCFHA-LVs at MOI = 1 allowed transduction of the hCD34+ cells within the MCs to levels equivalent to those of isolated CD34+ cells (Figure 2A vs Figure 1D).
In conclusion, RDTR/SCFHA-LVs allow a highly selective transduction of the very small subpopulation of hCD34+ cells in the MC fraction of CB in vitro.
RDTR/SCFHA-LVs promote targeted transduction and correction of hCD34+ cells in unfractionated BM from FA patients
FA is a rare genetic syndrome characterized by progressive BM failure. Gene therapy by infusion of FA-corrected autologous HSCs may offer a potential cure (for review, see Tolar et al10 ). However, the collection of hCD34+ cells in FA patients is challenging because of the reduced numbers of progenitor cells present in their BM or mobilized peripheral blood (PB).7-9 In addition, the FA genetic defect causes fragility of HSCs.26 For these reasons, it would be beneficial to minimize ex vivo handling of these cells by transducing hCD34+ cells directly in unfractionated FA BM samples. We first confirmed that the RDTR/SCFHA-LVs permitted hCD34+ transduction (MOI = 1) in healthy BM MCs equivalent to the levels obtained in CB MCs (compare supplemental Figure 1A-B and Figure 2A-B, respectively).
In 3 FA patients, the percentage of hCD34+ cells in the mononuclear BM cells was low compared with healthy donors (supplemental Table 1). RDTR/SCFHA-LVs transduced efficiently and specifically the hCD34+ cells in the BM of all patients, even in the absence of RetroNectin. They were highly superior for hCD34+-cell transduction over VSV-G/SCFHA-LVs and RDTR-LVs and VSV-G–LVs in the presence of rSCF (Figure 3A-C). VSV-G/SCFHA-LVs showed no preferential transduction of CD34+ cells because they transduced CD34+ and CD34− cells to the same extent in total FA BM. In contrast, 6 different RDTR/SCFHA-LV preparations allowed a highly preferential transduction of CD34+ cells compared with CD34− cells (Figure 3D). Multipotential differentiation was confirmed for transduced FA CD34+ cells by their differentiation into myeloid colonies (colony-forming cells or CFCs). Crucially, the percentage of GFP+ CFCs was equivalent to the percentage of GFP+hCD34+ cells from which the CFCs were derived (Figure 3E).
RDTR/SCFHA-LVs target gene transfer into hCD34+ cells in BM of FA patients. BM MCs of 3 different FA patients (FA-P1, FA-P2, and FA-P3 shown in panels A, B, and C, respectively) were incubated with 2 different vector preparations of RDTR/SCFHA-displaying or VSV-G/SCF–displaying LVs. As controls, the FA BM MCs were incubated with LVs displaying RDTR or VSV-G in the presence of rSCF (50 ng/mL); MOI = 10 was applied for all transductions. RetroNectin was added for all transductions except where indicated (n.d. indicates not done). At day 3 after transduction, cells were evaluated for CD34+ staining and GFP expression by FACS. (D) Transduction of total BM of a FA patient incubated with 6 different vector preparations of RDTR/SCFHA-displaying LVs or 2 different preparations of VSV-G/SCF–displaying LVs. At day 3 after transduction, cells were evaluated for hCD34+ cell-surface expression and GFP expression by FACS. (E) Comparison of transduction (GFP+) of hCD34+ cells 3 days after transduction and their derived CFCs 14 days after myeloid differentiation from total BM of FA-P3 to confirm stable transduction. (F) Total BM of a FANC-A patient was transduced with RDTR/SCF-displaying control vector encoding for GFP (SFFVEGFP) or a correcting vector encoding for FANCA and GFP (SFFVFAIEG). Total BM cells were transduced at MOI = 10. At day 3 of transduction, 1 × 105 total BM cells were cultured in methylcellulose that supported myeloid differentiation in the presence and absence of mitomycin C (10nM). GFP+ colonies per 1 × 105 seeded cells were scored.
RDTR/SCFHA-LVs target gene transfer into hCD34+ cells in BM of FA patients. BM MCs of 3 different FA patients (FA-P1, FA-P2, and FA-P3 shown in panels A, B, and C, respectively) were incubated with 2 different vector preparations of RDTR/SCFHA-displaying or VSV-G/SCF–displaying LVs. As controls, the FA BM MCs were incubated with LVs displaying RDTR or VSV-G in the presence of rSCF (50 ng/mL); MOI = 10 was applied for all transductions. RetroNectin was added for all transductions except where indicated (n.d. indicates not done). At day 3 after transduction, cells were evaluated for CD34+ staining and GFP expression by FACS. (D) Transduction of total BM of a FA patient incubated with 6 different vector preparations of RDTR/SCFHA-displaying LVs or 2 different preparations of VSV-G/SCF–displaying LVs. At day 3 after transduction, cells were evaluated for hCD34+ cell-surface expression and GFP expression by FACS. (E) Comparison of transduction (GFP+) of hCD34+ cells 3 days after transduction and their derived CFCs 14 days after myeloid differentiation from total BM of FA-P3 to confirm stable transduction. (F) Total BM of a FANC-A patient was transduced with RDTR/SCF-displaying control vector encoding for GFP (SFFVEGFP) or a correcting vector encoding for FANCA and GFP (SFFVFAIEG). Total BM cells were transduced at MOI = 10. At day 3 of transduction, 1 × 105 total BM cells were cultured in methylcellulose that supported myeloid differentiation in the presence and absence of mitomycin C (10nM). GFP+ colonies per 1 × 105 seeded cells were scored.
We also evaluated whether a RDTR/SCFHA-LV encoding a FANCA gene would allow correction of FA CD34+ cells. We transduced total BM cells from one FA-A patient with 2 different RDTR/SCFHA-LVs, one carrying EGFP under the control of the SFFV promoter (SFFVEGFP) and the other carrying the correcting FANCA gene driven by the SFFV promoter followed by an IRES-element driving the EGFP (SFFVFAIEG).
At MOI = 10, the GFP-encoding RDTR/SCFHA-LV selectively transduced 20% of the CD34+ cells in the total BM, whereas the FANCA-encoding RDTR/SCFHA LVs resulted in 40% selective transduction of the hCD34+ cells in the BM of the patients (data not shown).
Cells that regain FANCA expression become resistant to the mitomycin C, a classic DNA interstrand cross-linker highly toxic for FA cells. Indeed, BM CD34+ cells transduced with the FANCA-correcting vector resulted in mitomycin C–resistant CFC colonies, whereas the cells transduced with the GFP-expressing control vector yielded no CFC colonies (Figure 3F).
We conclude that very low doses of RDTR/SCFHA LVs confer efficient targeted FANC gene transfer to hCD34+ cells in unfractionated BM from FA patients, allowing them to bypass FA hCD34+ cell isolation. This is a procedure that would limit the collection of high amounts of functional HSCs and is a very significant step forward in the treatment of this disease.
RDTR/SCFHA-displaying vectors target transduction of CD34+ cells in total CB, mimicking in vivo–like administration
To mimic as closely as possible the in vivo setting for targeted gene transfer into hCD34+ cells, we performed transductions of fresh total CB containing active complement. Indeed, CB and adult serum inactivated VSV-G/SCF LVs but not RDTR/SCFHA LVs, as revealed by CD34+-cell transduction (supplemental Figure 2A). Because the ultimate goal is to inject this vector locally into the human BM (hBM) cavity, we compared the stability of the RDTR, RDTR/SCFHA-, VSV-G, and VSV-G/SCFHA-LVs against complement factors present in hBM aspirates. RDTR- and RDTR/SCF-LVs resisted BM complement inactivation whereas VSV-G and VSV-G/SCF LVs did not (supplemental Figure 2B).
Incubation of the vector with total CB allowed us to evaluate targeted gene transfer in hCD34+ cells, representing only 0.001% of cells in the blood. In addition, in this setting, the LVs are exposed to active complement in CB, an obstacle that will be encountered in vivo.
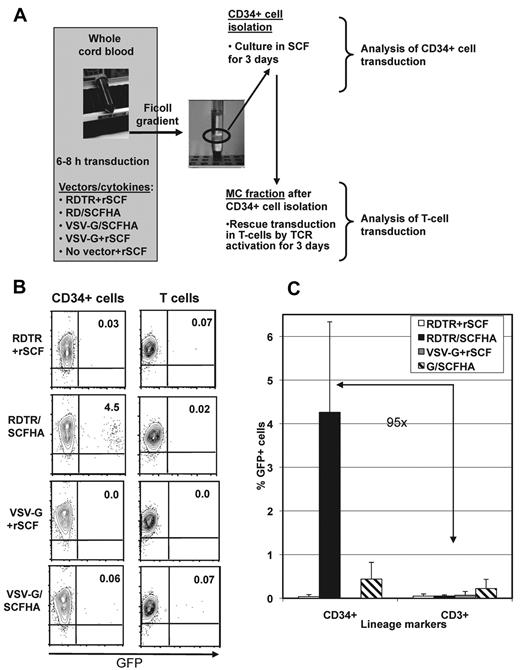
Total CB was incubated with RDTR- or VSV-G–LVs in the presence of rSCF or with RDTR/SCFHA- or VSV-G/SCFHA-LVs without adding exogenous cytokines. The experimental setup is outlined in Figure 4A. After 6-8 hours of incubation of the CB with the different vector pseudotypes, the hCD34+ cells were isolated and the residual MCs consisting mainly of T cells were cultured in the presence of anti-CD3/anti-CD28/rhIL-2 with the purpose of revealing potential off-target gene transfer in T cells. RDTR/SCFHA-LVs allowed a 10-fold higher level of transduction of hCD34+ cells (4.5%) than VSV-G/SCFHA-LVs (0.4%), whereas transductions with VSV-G- or RDTR-LVs plus rSCF were extremely inefficient (Figure 4B-C). VSV-G/SCFHA LVs showed consistently off-target T-cell transduction, resulting in only 1.8-fold selectivity for hCD34+cells. Considering that the blood contained 100-fold fewer hCD34+ cells than T cells, it is clear that RDTR/SCFHA-LVs selectively target transduction to hCD34+ cells with a 95-fold selectivity for CD34+ cells compared with T cells (Figure 4C).
RDTR/SCFHA-displaying LVs preferentially transduce hCD34+ cells in whole CB–containing active human complement. (A) Outline of the experimental setup. Total hCB-containing active complement was incubated with RDTR- or VSV-G–LVs in the presence of rSCF (50 ng/mL) or with vectors co-displaying the RDTR or VSV-G together with SCFHA glycoprotein without the addition of exogenous cytokines. Vectors were used at MOI = 0.001 calculated for the total number of WBCs and RBCs present in the blood sample. After 6-8 hours of incubation with the LVs, the CD34+ cells were isolated by positive selection and were cultured in medium containing 50 ng/mL of recombinant rSCF. After removal of the CD34+ cells, the residual MCs were cultured in RPMI supplemented with anti-CD3, anti-CD28, and rhIL-2. After 3 days of culture, transduction of very early progenitors (CD34+GFP+ cells) and T cells (CD3+GFP+ cells) in the MC fraction was analyzed by FACS. The dot blots are represented in panel B. The percentages of CD34+GFP+ cells (left) and GFP+ T cells (right) are indicated. The data are representative of 5 experiments. (C) Comparison of the transduction efficiencies of early progenitors (CD34+ cells) and T cells (CD3+ cells) in the whole CB sample.
RDTR/SCFHA-displaying LVs preferentially transduce hCD34+ cells in whole CB–containing active human complement. (A) Outline of the experimental setup. Total hCB-containing active complement was incubated with RDTR- or VSV-G–LVs in the presence of rSCF (50 ng/mL) or with vectors co-displaying the RDTR or VSV-G together with SCFHA glycoprotein without the addition of exogenous cytokines. Vectors were used at MOI = 0.001 calculated for the total number of WBCs and RBCs present in the blood sample. After 6-8 hours of incubation with the LVs, the CD34+ cells were isolated by positive selection and were cultured in medium containing 50 ng/mL of recombinant rSCF. After removal of the CD34+ cells, the residual MCs were cultured in RPMI supplemented with anti-CD3, anti-CD28, and rhIL-2. After 3 days of culture, transduction of very early progenitors (CD34+GFP+ cells) and T cells (CD3+GFP+ cells) in the MC fraction was analyzed by FACS. The dot blots are represented in panel B. The percentages of CD34+GFP+ cells (left) and GFP+ T cells (right) are indicated. The data are representative of 5 experiments. (C) Comparison of the transduction efficiencies of early progenitors (CD34+ cells) and T cells (CD3+ cells) in the whole CB sample.
In conclusion, RDTR/SCFHA-LVs promote targeted gene transfer into hCD34+ progenitor cells in the in vivo–like conditions encountered in a whole CB sample.
RDTR/SCFHA-engineered LVs transduce hHSCs capable of long-term immunodeficient mice reconstitution
Having previously obtained reproducibly high reconstitution of the Balbc rag2−/−γc−/− mice with LV-transduced CB hCD34+ cells,27 we chose this model to evaluate the long-term reconstitution capacity of RDTR/SCFHA-LV–transduced hCD34+ cells in vivo. In Figure 5A, a representative mouse is shown that was reconstituted 16 weeks before with RDTR/SCFHA-LV–transduced hCD34+ cells (MOI = 1). As shown in Figure 5A, 7% of BM hCD45+ cells expressed GFP. Detailed phenotyping showed that very early progenitor CD34+ cells, as well as B-cell CD19+ progenitors and myeloid CD13+ progenitors, were transduced to the same extent (Table 1 and Figure 5A). This was also found for T and B cells and monocytes in the spleen (Figure 5B). Transduced human T and B cells were detected in the blood and the different thymic subpopulations (Figure 5C-D). We confirmed this transduction profile for 6 mice reconstituted with RDTR/SCFHA-LV–transduced hCD34+ cells (Table 1). Note that incubation of hCD34+ cells with both high (MOI = 10) and low (MOI = 1) vector doses resulted in transduction of myeloid (CD14+ and CD13+ cells) and lymphoid lineages (CD3+ and CD19+ cells).
Multilineage reconstitution of Balbc rag2−/−γc−/− mice with RDTR/SCFHA-LV–transduced hCD34+ cells. (A) FACS analysis of multilineage engraftment in BM of a Balbc rag2−/−γc−/− mouse reconstituted for 16 weeks with RDTR/SCFHA-LV–transduced CB CD34+ cells. The top right quadrants show the GFP+ cells within the early progenitors (hCD34+), B cells (hCD19+), myeloid progenitors (hCD13+), and monocytes (hCD14+) in the human graft (hCD45+ cells). FACS analysis of multilineage engraftment of the RDTR/SCFHA-LV–transduced CB CD34+ cells in the spleen and in PB is shown in panels B and C, respectively. Panel D shows the transduced cells in the thymic subpopulation: SP-4 (CD4+CD8−), SP-8 (CD4−CD8+), and DP (CD4+CD8+). The FACS profiles are representative of 6 experiments listed in Table 1.
Multilineage reconstitution of Balbc rag2−/−γc−/− mice with RDTR/SCFHA-LV–transduced hCD34+ cells. (A) FACS analysis of multilineage engraftment in BM of a Balbc rag2−/−γc−/− mouse reconstituted for 16 weeks with RDTR/SCFHA-LV–transduced CB CD34+ cells. The top right quadrants show the GFP+ cells within the early progenitors (hCD34+), B cells (hCD19+), myeloid progenitors (hCD13+), and monocytes (hCD14+) in the human graft (hCD45+ cells). FACS analysis of multilineage engraftment of the RDTR/SCFHA-LV–transduced CB CD34+ cells in the spleen and in PB is shown in panels B and C, respectively. Panel D shows the transduced cells in the thymic subpopulation: SP-4 (CD4+CD8−), SP-8 (CD4−CD8+), and DP (CD4+CD8+). The FACS profiles are representative of 6 experiments listed in Table 1.
RDTR/SCFHA-LV–transduced hCD34+-cell engraftment and multilineage differentiation
| Mice . | MOI . | BM, % . | Spleen, % . | Thy, % . | PB, % . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD45+/CD45+ GFP+ . | CD34+ GFP+ . | CD13+ GFP+ . | CD14+ GFP+ . | CD19+ GFP+ . | CD3+ GFP+ . | CD14+ GFP+ . | CD19+ GFP+ . | CD45+ GFP+ . | CD19+ GFP+ . | CD3+ GFP+ . | ||
| 1 | 10 | 57/37 | 38 | 40 | 29 | 33 | 10 | 29 | 38 | 23 | 35 | 30 |
| 2 | 10 | 51/37 | 33 | 46 | 39 | 20 | 80 | 37 | 40 | 34 | 50 | 20 |
| 3 | 1 | 47/7 | 10.5 | 6 | 9.5 | 5 | 9 | 10 | 7 | 7 | 7.4 | 10 |
| 4 | 1 | 50/5.5 | 7 | 6 | 6.5 | 5.1 | 7.6 | 5.5 | 6.1 | 6 | 5 | 3.5 |
| 5 | 1 | 15/4 | 4 | 4.5 | 5 | 3 | 1 | 10 | 7.5 | 6 | 11 | ND |
| 6 | 1 | 8/10 | 10 | 6.1 | 8 | 8.5 | 2 | 5 | 7.2 | 9.3 | ND | ND |
| Mice . | MOI . | BM, % . | Spleen, % . | Thy, % . | PB, % . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD45+/CD45+ GFP+ . | CD34+ GFP+ . | CD13+ GFP+ . | CD14+ GFP+ . | CD19+ GFP+ . | CD3+ GFP+ . | CD14+ GFP+ . | CD19+ GFP+ . | CD45+ GFP+ . | CD19+ GFP+ . | CD3+ GFP+ . | ||
| 1 | 10 | 57/37 | 38 | 40 | 29 | 33 | 10 | 29 | 38 | 23 | 35 | 30 |
| 2 | 10 | 51/37 | 33 | 46 | 39 | 20 | 80 | 37 | 40 | 34 | 50 | 20 |
| 3 | 1 | 47/7 | 10.5 | 6 | 9.5 | 5 | 9 | 10 | 7 | 7 | 7.4 | 10 |
| 4 | 1 | 50/5.5 | 7 | 6 | 6.5 | 5.1 | 7.6 | 5.5 | 6.1 | 6 | 5 | 3.5 |
| 5 | 1 | 15/4 | 4 | 4.5 | 5 | 3 | 1 | 10 | 7.5 | 6 | 11 | ND |
| 6 | 1 | 8/10 | 10 | 6.1 | 8 | 8.5 | 2 | 5 | 7.2 | 9.3 | ND | ND |
Two- to 4-day-old newborn BALB/c Rag2−/−γc−/− mice were subjected to a sublethal irradiation of 2 × 1.5 Gy. hCB-CD34+ cells (2 × 105) transduced with RDTR/SCFHA-LVs at the indicated MOI were injected intrahepatically into the newborns. After 12 to 16 weeks of reconstitution, the humanized mice were killed and the BM, spleens, thymi, and PB were harvested and assessed for levels of human engraftment and GFP-expressing human cells. Multilineage engraftment was demonstrated by Lin+ markers as indicated, and for each lineage, the percentage of EGFP+ cells was determined by FACS. Independent experiments were performed with different CD34+ CB samples and different preparations of each vector.
ND indicates no human cell engraftment detected.
Overall, these data strongly suggest that RDTR/SCFHA LVs allow for the transduction of very early progenitor repopulating cells—so-called hHSCs—able to differentiate into all of the different lineages in immunodeficient mice.
RDTR/SCFHA-LVs allow direct in vivo gene transfer into hHSCs
Because RDTR/SCFHA LVs selectively transduced hCD34+ cells in unfractionated BM (Figure 3D), we evaluated the capacity of these LVs to allow in vivo gene transfer into hHSCs using humanized Balbc rag2−/−γc−/− mice in which hCD34+ CB cells were transplanted 13 weeks before the intrafemoral LV inoculation. After 13 weeks of hCD34+-cell engraftment in these mice, we detected 5%-46% of hCD45+ cells in their BM (Table 2). Importantly, 5%-10% of the human cells expressed the hCD34+ surface marker, representing the in vivo target cells in the BM. We injected 2 × 105-1 × 106 infectious units of the RDTR/SCFHA LVs or the RDTR LVs locally into both femurs of the humanized mice (MOI = 0.01-0.05).
Evaluation of lentivectors for in vivo transduction of HSCs in humanized mice
| Mice . | Reconstitution, % hCD45+cells* . | BM, % GFP+ cells/total Lin+ cells† . | Thymus, % GFP+ cells† . | Spleen, % GFP+ cells† . | PB, % GFP+ cells† . | Vector titer, IU/mL . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BM . | Thymus . | Spleen . | PB . | h45+ . | h34+ . | h13+ . | h19+ . | h45+ . | h45+ . | h45+ . | |||
| RDTR/SCFHA 1 | 7 | 80 | 6 | F1 | 2.5 | 6.6 | 1 | 0.4 | 0 | 0 | 0 | 4E8 | |
| F2 | 2.3 | 6.5 | 6.6 | 0.3 | |||||||||
| RDTR/SCFHA 2 | 40 | 85 | 56 | 2 | F1 | 2.5 | 3.3 | 5 | 0.7 | 0 | 0.05 | 0 | 9E7 |
| F2 | 3.8 | 4 | 7.3 | 0.5 | |||||||||
| RDTR/SCFHA 3 | 18 | 98 | 47 | 7 | F1 | 2.2 | 4.3 | 6.6 | 0.06 | 0 | 0.006 | 0 | 1E8 |
| F2 | |||||||||||||
| RDTR/SCFHA 4 | 46 | 93 | 30 | 3 | F1 | 2.1 | 2.6 | 6.1 | 0.8 | 0 | 0.04 | 0 | 2.9E7 |
| F2 | 1.7 | 3 | 4.2 | 0.7 | |||||||||
| RDTR/SCFHA 5 | 40 | 99 | 15 | 5 | F1 | 1.9 | 2.2 | 6.8 | 0.7 | 0 | 0.06 | 0 | 4E7 |
| F2 | 2.8 | 4.2 | 4.5 | 0.7 | |||||||||
| RDTR/SCFHA 6 | 19 | 94 | 36 | 30 | F1 | 2 | 2.6 | 2.5 | 0.03 | 0 | 0.03 | 0 | 5E7 |
| F2 | 2.8 | 2.1 | 6.1 | 0.04 | |||||||||
| RDTR/SCFHA 7 | 6 | 97 | 5 | 1 | F1 | 1.2 | 4 | 2.9 | 0.8 | 0 | 0 | 0 | 2.9E7 |
| F2 | |||||||||||||
| Means | 2.3 | 3.8 | 4.9 | 0.4 | 0 | 0.02 | 0 | ||||||
| SD (±) | 0.6 | 1.5 | 1.9 | 0.3 | 0 | 0.02 | 0 | ||||||
| RDTR 1 | 10 | 91 | 30 | 4 | F1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.3E8 |
| F2 | 0 | 0 | 0 | 0 | |||||||||
| RDTR 2 | 7 | 90 | 5 | 6 | F1 | 0.05 | 0 | 0 | 0.05 | 0 | 0 | 0 | 2E8 |
| F2 | 0.4 | 0 | 0 | 0.05 | |||||||||
| RDTR 3 | 15 | 93 | 20 | 11 | F1 | 0.6 | 0 | 0 | 0.6 | 0 | 0 | 0 | 2E8 |
| F2 | 0.8 | 0 | 0 | 0.8 | |||||||||
| RDTR 4 | 7 | 97 | 12 | 4 | F1 | 0.1 | 0 | 0 | 0.3 | 0 | 0 | 0 | 4E7 |
| F2 | 0.2 | 0 | 0 | 0.4 | |||||||||
| Means | 0.3 | 0 | 0 | 0.3 | 0 | 0 | 0 | ||||||
| SD (±) | 0.3 | 0 | 0 | 0.3 | 0 | 0 | 0 | ||||||
| RDTR 1+ SCFHA 1 | 21 | 92 | 18 | 10 | F1 | 0.04 | 0 | 0.01 | 0.03 | 0 | 0 | 0 | 1.3E8 |
| F2 | 0.01 | 0 | 0.00 | 0.01 | |||||||||
| RDTR 2+ SCFHA 2 | 17 | 95 | 11 | 4 | F1 | 0.02 | 0 | 0 | 0.03 | 0 | 0 | 0 | 2E8 |
| F2 | 0.2 | 0 | 0 | 0.25 | |||||||||
| RDTR 3+ SCFHA 3 | 5 | 86 | 7 | F1 | 0.0 | 0 | 0 | 0 | 0 | 0 | 0 | 4E7 | |
| F2 | 0.2 | 0 | 0 | 0.3 | |||||||||
| Mice . | Reconstitution, % hCD45+cells* . | BM, % GFP+ cells/total Lin+ cells† . | Thymus, % GFP+ cells† . | Spleen, % GFP+ cells† . | PB, % GFP+ cells† . | Vector titer, IU/mL . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BM . | Thymus . | Spleen . | PB . | h45+ . | h34+ . | h13+ . | h19+ . | h45+ . | h45+ . | h45+ . | |||
| RDTR/SCFHA 1 | 7 | 80 | 6 | F1 | 2.5 | 6.6 | 1 | 0.4 | 0 | 0 | 0 | 4E8 | |
| F2 | 2.3 | 6.5 | 6.6 | 0.3 | |||||||||
| RDTR/SCFHA 2 | 40 | 85 | 56 | 2 | F1 | 2.5 | 3.3 | 5 | 0.7 | 0 | 0.05 | 0 | 9E7 |
| F2 | 3.8 | 4 | 7.3 | 0.5 | |||||||||
| RDTR/SCFHA 3 | 18 | 98 | 47 | 7 | F1 | 2.2 | 4.3 | 6.6 | 0.06 | 0 | 0.006 | 0 | 1E8 |
| F2 | |||||||||||||
| RDTR/SCFHA 4 | 46 | 93 | 30 | 3 | F1 | 2.1 | 2.6 | 6.1 | 0.8 | 0 | 0.04 | 0 | 2.9E7 |
| F2 | 1.7 | 3 | 4.2 | 0.7 | |||||||||
| RDTR/SCFHA 5 | 40 | 99 | 15 | 5 | F1 | 1.9 | 2.2 | 6.8 | 0.7 | 0 | 0.06 | 0 | 4E7 |
| F2 | 2.8 | 4.2 | 4.5 | 0.7 | |||||||||
| RDTR/SCFHA 6 | 19 | 94 | 36 | 30 | F1 | 2 | 2.6 | 2.5 | 0.03 | 0 | 0.03 | 0 | 5E7 |
| F2 | 2.8 | 2.1 | 6.1 | 0.04 | |||||||||
| RDTR/SCFHA 7 | 6 | 97 | 5 | 1 | F1 | 1.2 | 4 | 2.9 | 0.8 | 0 | 0 | 0 | 2.9E7 |
| F2 | |||||||||||||
| Means | 2.3 | 3.8 | 4.9 | 0.4 | 0 | 0.02 | 0 | ||||||
| SD (±) | 0.6 | 1.5 | 1.9 | 0.3 | 0 | 0.02 | 0 | ||||||
| RDTR 1 | 10 | 91 | 30 | 4 | F1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.3E8 |
| F2 | 0 | 0 | 0 | 0 | |||||||||
| RDTR 2 | 7 | 90 | 5 | 6 | F1 | 0.05 | 0 | 0 | 0.05 | 0 | 0 | 0 | 2E8 |
| F2 | 0.4 | 0 | 0 | 0.05 | |||||||||
| RDTR 3 | 15 | 93 | 20 | 11 | F1 | 0.6 | 0 | 0 | 0.6 | 0 | 0 | 0 | 2E8 |
| F2 | 0.8 | 0 | 0 | 0.8 | |||||||||
| RDTR 4 | 7 | 97 | 12 | 4 | F1 | 0.1 | 0 | 0 | 0.3 | 0 | 0 | 0 | 4E7 |
| F2 | 0.2 | 0 | 0 | 0.4 | |||||||||
| Means | 0.3 | 0 | 0 | 0.3 | 0 | 0 | 0 | ||||||
| SD (±) | 0.3 | 0 | 0 | 0.3 | 0 | 0 | 0 | ||||||
| RDTR 1+ SCFHA 1 | 21 | 92 | 18 | 10 | F1 | 0.04 | 0 | 0.01 | 0.03 | 0 | 0 | 0 | 1.3E8 |
| F2 | 0.01 | 0 | 0.00 | 0.01 | |||||||||
| RDTR 2+ SCFHA 2 | 17 | 95 | 11 | 4 | F1 | 0.02 | 0 | 0 | 0.03 | 0 | 0 | 0 | 2E8 |
| F2 | 0.2 | 0 | 0 | 0.25 | |||||||||
| RDTR 3+ SCFHA 3 | 5 | 86 | 7 | F1 | 0.0 | 0 | 0 | 0 | 0 | 0 | 0 | 4E7 | |
| F2 | 0.2 | 0 | 0 | 0.3 | |||||||||
Two- to 4-day-old newborn BALB/c Rag2−/−γc−/− mice were subjected to a sublethal irradiation of 2 × 1.5 Gy. 2 × 105 hCB CD34+ cells were injected intrahepatically into the newborns. After 13 weeks of reconstitution, the humanized mice were injected into the right (F1) and the left (F2) femur with 10 μL of the indicated vector stock. The animals were killed 2 weeks after intrafemoral injection of the vectors, and the BM, spleens, thymi, and PB were harvested and assessed for levels of human cell engraftment. Multilineage engraftment was demonstrated by Lin+ markers as indicated, and the percentage of EGFP+ cells was determined by FACS. Independent experiments were performed with different CD34+ CB samples and different preparations of each vector. Vector titers (IU/mL) were determined on 293T cells
Percentage of human cells (hCD45+ cells) per total number of human and mouse mononucleated cells in each hematopoietic tissue.
Percentage of GFP+ cells per total Lin+ cells (ie, % GFP+CD13+ cells/total CD13+ cells).
In the flushed BM of the mice, 2 weeks after injection with the RDTR/SCFHA-LVs, we revealed on average 2.3% transduction of the human cells (hCD45+) that had colonized the mice BM (Table 2 and Figure 6A). Interestingly, a transduction of 2.1%-6.6% of early human progenitors (CD34+) and 2.9%-7.3% of total myeloid progenitors (CD13+) in the BM was detected (Table 2 and Figure 6B). Nontarget cells, such as monocytes (CD14+) and pre- and pro-B cells (CD19+), were transduced to a much lower extent in the BM (Table 2). In sharp contrast, RDTR- or RDTR- plus SCFHA- LVs combined intra-BM injection resulted in low transduction of hBM cells (Table 2). The few cells transduced in the BM were already committed to the B-cell lineage emphasizing the requirement for co-display of RDTR and SCF on the same LV to obtain efficient hCD34+-cell transduction in the BM in vivo (Table 2).
RDTR/SCFHA-displaying LVs allow in vivo gene transfer into immature hCD34+ cells in humanized mice. Newborn Balbc rag2−/−γc−/− mice received sublethal irradiation (2 × 1.5 Gy) and then 2 × 105 hCD34+ CB cells were injected into the fetal liver at day 2 to 4 after birth. After 13 weeks of reconstitution, RDTR/SCFHA-LVs were injected into both femurs of the humanized mice. Two to 3 weeks after injection, the mice were killed and the transduction of the different cell lineages in the BM was analyzed (A-D). The percentage of transduced human cells (hCD45+GFP+) is shown for 3 different injected animals listed in Table 2 (A). In panel B, transduction (GFP+) of immature progenitors (hCD34+), myeloid progenitors (hCD13+), monocytes (hCD14+), and pre- and pro-B cells (hCD19+) in the BM was analyzed by FACS. (C) Transduction in different BM subpopulations by CD34/CD19 double staining. Transduction is shown for very immature, nonlineage-committed CD34+ cells (CD34+CD19−), for pre- and pro-B cells (CD34+CD19+) and further differentiated B cells (CD34−CD19+). In panel D, transduction of immature B cells (CD19+CD20−) and of mature B cells (CD19+CD20+) are presented in the dot blots.
RDTR/SCFHA-displaying LVs allow in vivo gene transfer into immature hCD34+ cells in humanized mice. Newborn Balbc rag2−/−γc−/− mice received sublethal irradiation (2 × 1.5 Gy) and then 2 × 105 hCD34+ CB cells were injected into the fetal liver at day 2 to 4 after birth. After 13 weeks of reconstitution, RDTR/SCFHA-LVs were injected into both femurs of the humanized mice. Two to 3 weeks after injection, the mice were killed and the transduction of the different cell lineages in the BM was analyzed (A-D). The percentage of transduced human cells (hCD45+GFP+) is shown for 3 different injected animals listed in Table 2 (A). In panel B, transduction (GFP+) of immature progenitors (hCD34+), myeloid progenitors (hCD13+), monocytes (hCD14+), and pre- and pro-B cells (hCD19+) in the BM was analyzed by FACS. (C) Transduction in different BM subpopulations by CD34/CD19 double staining. Transduction is shown for very immature, nonlineage-committed CD34+ cells (CD34+CD19−), for pre- and pro-B cells (CD34+CD19+) and further differentiated B cells (CD34−CD19+). In panel D, transduction of immature B cells (CD19+CD20−) and of mature B cells (CD19+CD20+) are presented in the dot blots.
To confirm that RDTR/SCFHA-LVs transduced immature hCD34+ cells in vivo, a CD34+/CD19+ double staining was performed. Of utmost importance, a 10-fold higher transduction of hCD34+CD19− cells not yet showing B-cell commitment was detected repeatedly compared with the early B-cell progenitors (CD34+CD19+) and more committed B cells (CD34−CD19+; Figure 6C). This was in accordance with the fact that more mature B cells in the BM (CD20+CD19+) were minimally transduced (Figure 6D).
Two weeks after injection, we verified in vivo escape of the vectors into the circulation. We did not detect GFP+ thymocytes (supplemental Figure 3A), nor transduction of human B and T cells in the blood of these intra-BM–injected mice (supplemental Figure 3A); we also detected very low levels of transduced B cells in the spleen (Table 2 and supplemental Figure 3C).
To confirm the data shown in Table 2, a similar experiment was conducted in which animals were analyzed 6-8 weeks after in vivo inoculation of the LVs. An identical hCD34+-cell transduction profile was revealed 6-8 weeks after RDTR/SCFHA-LV injection, with a slight transduction level increase in more differentiated cells such as mature B cells and myeloid cells compared with 2 weeks after injection of vector (Table 3 vs Table 2). Remarkably, isolation of hCD34+ BM cells from these 6- to 8-week RDTR/SCFHA-LV–injected mice and their differentiation into myeloid cell colonies (CFUs) revealed levels of CFU transduction that were equivalent to the transduction levels of the hCD34+ BM cells in vivo from which they were derived (Table 3). Furthermore, after myeloid differentiation, we detected GFP+-CFU-M, CFU-G, BFU-E, CFU-GM, and CFU-GEMM colonies and the transduction rates in the different colony types were very similar (supplemental Figure 4). The fact that we detected transduced CFU-GM and CFU-GMME mixed colonies strongly indicated that the initial target cells were very primitive candidate HSCs (supplemental Figure 4).
RDTR/SCFHA-LV long-term in vivo transduction of hHSCs in humanized mice
| Mice . | Reconstitution, % hCD45+ cells* . | BM, % GFP+ cells/total Lin+ cells† . | Thymus, % GFP+ cells† . | Spleen, % GFP+ cells† . | PB, % GFP+ cells† . | CFCs, % GFP+ cells‡ . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BM . | Thy . | Spl . | PB . | h45 . | h34 . | h13 . | h19 . | h45 . | h45 . | h45 . | |||
| RDTR/SCFHA 1 | 18 | 98 | 24 | 9 | IF1 + 2 | 4 | 3.9 | 4 | 1.7 | 0 | 0.1 | 0 | 4.6 |
| RDTR/SCFHA 2 | 25 | 97 | 21 | 5 | IF1 + 2 | 2.4 | 3.6 | 3 | 0.2 | 0 | 0 | 0 | ND |
| RDTR/SCFHA 3 | 10 | 98.3 | 20 | 1 | IF1 + 2 | 3.4 | 5.7 | 7.1 | 1.1 | 0 | 0.3 | 0 | 4.3 |
| RDTR/SCFHA 4 | 5 | 97 | 10 | 2 | IF1 + 2 | 5.5 | 4 | 5.4 | 0.8 | 0 | 0.4 | 0 | 5.5 |
| RDTR/SCFHA 5 | 16 | 91 | 49 | 6 | IF1 + 2 | 3.7 | 5.5 | 5.3 | 1.6 | 0 | 0.4 | 0 | 5.3 |
| RDTR/SCFHA 6 | 7 | 98 | 38 | ND | IF1 + 2 | 3.4 | 4.9 | 4.1 | 1.4 | 0 | 0.1 | 3.8 | |
| RDTR/SCFHA 7 | 5 | 99 | 42 | 2 | IF1 + 2 | 3 | 6.4 | 8.9 | 0.5 | 0 | 0.03 | 0 | 5.8 |
| Mice . | Reconstitution, % hCD45+ cells* . | BM, % GFP+ cells/total Lin+ cells† . | Thymus, % GFP+ cells† . | Spleen, % GFP+ cells† . | PB, % GFP+ cells† . | CFCs, % GFP+ cells‡ . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BM . | Thy . | Spl . | PB . | h45 . | h34 . | h13 . | h19 . | h45 . | h45 . | h45 . | |||
| RDTR/SCFHA 1 | 18 | 98 | 24 | 9 | IF1 + 2 | 4 | 3.9 | 4 | 1.7 | 0 | 0.1 | 0 | 4.6 |
| RDTR/SCFHA 2 | 25 | 97 | 21 | 5 | IF1 + 2 | 2.4 | 3.6 | 3 | 0.2 | 0 | 0 | 0 | ND |
| RDTR/SCFHA 3 | 10 | 98.3 | 20 | 1 | IF1 + 2 | 3.4 | 5.7 | 7.1 | 1.1 | 0 | 0.3 | 0 | 4.3 |
| RDTR/SCFHA 4 | 5 | 97 | 10 | 2 | IF1 + 2 | 5.5 | 4 | 5.4 | 0.8 | 0 | 0.4 | 0 | 5.5 |
| RDTR/SCFHA 5 | 16 | 91 | 49 | 6 | IF1 + 2 | 3.7 | 5.5 | 5.3 | 1.6 | 0 | 0.4 | 0 | 5.3 |
| RDTR/SCFHA 6 | 7 | 98 | 38 | ND | IF1 + 2 | 3.4 | 4.9 | 4.1 | 1.4 | 0 | 0.1 | 3.8 | |
| RDTR/SCFHA 7 | 5 | 99 | 42 | 2 | IF1 + 2 | 3 | 6.4 | 8.9 | 0.5 | 0 | 0.03 | 0 | 5.8 |
Two- to 4-day-old newborn BALB/c Rag2−/−γc−/− mice were subjected to a sublethal irradiation of 2 × 1.5 Gy. 2 × 105 hCB-CD34+ cells were injected intrahepatically into the newborns. After 9 weeks of reconstitution, the humanized mice were injected into both femurs with 10 μL of the RDTR/SCFHA-LVs (5E7–1E8 IU/mL). The animals were killed 6-8 weeks after intrafemoral injection of the vectors and the BM, spleens, thymi, and PB were harvested and assessed for levels of human engraftment and GFP-expressing human cells. Independent experiments were performed with different CD34+ CB samples and different preparations of each vector.
ND indicates not done.
Percentage of human cells (hCD45+ cells) per total human and mouse mononucleated cells in each hematopoietic tissue.
Percentages of GFP+ cells per total Lin+ cells (ie, the percentage of GFP+CD13+ cells/total CD13+ cells).
Six to 8 weeks after RDTR/SCFHA-LV intrafemoral injection into these humanized mice, hCD34+ cells were isolated from part of the humanized BM by magnetic cell isolation (Miltenyi Biotec) and then seeded in methyl cellulose to induce myeloid differentiation and to score the percentage of GFP+ CFCs.
In summary, local administration of low doses of RD/SCF-LVs into the BM of humanized mice resulted in a highly selective long-term transduction of Lin− hCD34+ cells in vivo.
Discussion
The novel RDTR/SCFHA-LVs reported herein allowed targeted gene transfer in very immature hCD34+ cells in the mononuclear fraction of human CB and BM. Moreover, in complete CB-retaining active complement, these RDTR/SCFHA-LVs transduced hCD34+ cells at low vector doses with a 95-fold selectivity compared with T cells (Figure 3). Most importantly, administration of low doses of RDTR/SCFHA-LVs into the BM of humanized mice resulted in a selective long-term transduction of human c-Kit+CD34+Lin− HSCs in vivo.
The choice of targeting molecule on hHSCs is not evident because as yet, no exclusive phenotypic marker specific for hHSC has been identified. We opted for presentation of SCF at the LV surface, allowing specific binding to c-Kit+CD34+ cells. We coexpressed RD114 glycoprotein as an “entry/fusion” glycoprotein because its receptor is present at high levels on hCD34+ cells and because RD114 glycoprotein LVs efficiently transduce HSCs.18,19 Insertion of a ligand such as SCF in the influenza HA allowed excellent functional presentation on the LV particles, but completely compromised the vector-cell fusion capacity of the HA glycoprotein. Therefore, SCFHA can bind to the c-Kit receptor but does not efficiently fuse with the target cell. Indeed, the titer of SCFHA-LVs was close to background (data not shown). This is a common phenomenon seen after ligand insertion into many retroviral envelope glycoproteins (for review, see Frecha et al28 ), indicating that the SCFHA glycoprotein in RDTR/SCFHA-LVs serves mainly to target the vector specifically to hCD34+c-Kit+ primary cells, allowing specific binding/stimulation through the c-Kit-receptors on CD34+ cells, whereas the RDTR glycoprotein allows vector-cell fusion. It will be very interesting to unravel the fate of these particles after cell entry. For in vivo applications, the resistance of the RDTR-LVs to degradation by human complement factors present in the hCB or hBM cavity is of utmost importance. In agreement with our results, a correcting RDTR vector has been injected in vivo into a canine X-SCID model, and an impressive correction of T-cell numbers and function was observed.29 These data indicate that in vivo, RDTR-LVs are resistant to canine complement. Moreover, DePolo et al showed a correlation between the half-life of a vector in vivo and its in vitro resistance to complement.30 In addition, VSV-G–LVs could bind to erythrocytes and thus be lost for hCD34+-cell transduction in vivo. Blocking the c-Kit-receptor confirmed that RDTR/SCFHA-LVs target transduction to c-Kit+hCD34+ cells. Accordingly, this LV showed very low gene transfer in off-target blood cells in vivo. Prestimulated T cells were ex vivo transduced by RDTR-LVs, but only in the presence of RetroNectin.17 In contrast, resting human T cells, which represent the majority of T cells in vivo, are poorly transduced by high doses of RDTR-LVs, whereas VSV-G–LVs used at high vector doses caused significant transduction (data not shown). As far as biosafety is concerned, the RDTR/SCFHA-LVs resulted in low copy integration, thus reducing the risk of genotoxicity (supplemental Table 2). Froelich et al incorporated both membrane-bound-SCF and a Sindbis-glycoprotein for vector-cell fusion at the surface of an LV, but the efficiency of these vectors for hCD34+-cell transduction was not tested.31 Another group used a Sindbis glycoprotein in which they incorporated the ZZ-domain of protein A conjugated with anti-CD34 Ab, which transduced hCD34+ cells. However, it is difficult to predict how these LVs will behave in vivo.32 One study performed intrafemoral injection of murine retroviral vectors into Jak-3 knockout mice.33 That study, however, used 5-fluorouracil preconditioning, which induces cycling and possibly differentiation of the very primitive early progenitors. In the present study, we did not use any preconditioning and the vector itself only presents SCF at the surface to maximize conservation of the HSC character. Only one study used intra-BM injection of LVs into mice, and this resulted in the transduction of mouse HSCs.34 However, a comparison with our data is not justified because that study used normal immunocompetent mice and we used human immune system mice. Our RDTR/SCF-LVs have a specific preference for human cells and cannot transduce mouse HSCs because the RD114 receptor ASCT-2 is not recognized on mouse cells.35 Human germ cells do not appear to express ASCT-2.36 Moreover, Ting-Deravin29 performed IV injection of concentrated RD114 glycoprotein-vector into X-SCID dogs and did not detect any transgene in DNA from sperm, although RD114 glycoprotein vectors can recognize canine ASCT-2. Taking both reports into account, it is highly unlikely that in vivo gene transfer with our vector will result in germline transmission of a therapeutic gene. However, to properly address the question of off-target transduction in vivo, RDTR/SCFHA-LV injection into primates will be needed.
Several inherited diseases are currently being treated or are being considered for treatment by ex vivo gene therapy, including adenosine deaminase–deficient SCID,1 X-SCID,2,37 Wiskott-Aldrich syndrome,38 FA,8,39,40 and β-thalassemia.41,42 All of these diseases are characterized by an in vivo selective advantage of corrected cells, and therefore they would clearly benefit from our in vivo LV-mediated HSC correction strategy. In FA, the genetic defect limits the number of HSCs in the BM of the patient and renders the hCD34+ cells fragile during in vitro manipulation (for review, see Tolar et al10 ). Therefore, transducing FA HSCs in total BM ex vivo (or even in vivo) without in vitro manipulation and transplantation of the corrected FA HSCs would improve the efficacy of FA gene therapy. In the present study, we have demonstrated that RDTR/SCFHA-LVs target gene transfer to hCD34+ cells in unfractionated BM of FA patients. Consistent with data obtained from mosaic patients,43-45 it is expected that genetically corrected HSCs will develop a marked proliferation advantage over uncorrected cells.43 It is therefore expected that the proportion of uncorrected versus genetically correct HSCs may progressively increase, limiting the risk of leukemia development.10,46 GALVTR-LVs allowed 25% FA progenitor-cell transduction.8 However, a prestimulation of the BM MCs with SCF + TPO + Flk-3L was performed before incubation with GALVTR-LVs preloaded on RetroNectin. We stimulated the hCD34+ cells only with SCF to conserve the HSC phenotype, and vectors and SCF were added simultaneously. Therefore, the 2 studies cannot be compared. The choice of RDTR over GALVTR glycoproteins for co-display with cytokines on LVs was obvious, because GALVTR-LVs cannot be efficiently concentrated, whereas RDTR LVs can17 and scale-up of GALVTR-LV production proved to be difficult.10 In other diseases, such as hemophilia, the clinical benefit of gene therapy relies on the bystander effect produced by the secretion of missing factors from corrected HSCs and their progeny.47 Therefore, all of these diseases would profit from this in vivo LV transduction strategy because patients should experience clinical benefit even if few HSCs are corrected.
The RDTR/SCFHA-LVs might also be used ex vivo to combine a CB transplantation with a gene therapy approach, because CB transplantations are currently an important alternative to BM transplantations.48 RDTR/SCFHA-LVs transduced the CB hCD34+ cell fraction at very low vector doses with a 95-fold selectivity compared with T cells (Figure 4C). The finding that RDTR/SCFHA-LVs allow highly specific transduction of the hCD34+ cells in CB might permit the augmentation of the number of corrected cells in vivo by co-introduction of positive selection markers, and this is currently under investigation.49
In summary, this new generation of HSC-targeted LVs should simplify and improve current gene therapy protocols through transduction of primitive HSCs directly in the BM of patients with genetic diseases. In the future, this new tool could replace all ex vivo handling and allow for the correction of multiple hematopoietic diseases via a direct intra-BM inoculum in vivo.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Mamoro Ito and Taconic (Central Institute for Experimental Animals, Kawasaki, Japan) for sharing the Balbc, rag2−/−γc−/− immunodeficient mice with us and the staff of the animal care facility (Le Plateau de Biologie Expérimental de la Souris) at the École Normale Supérieure de Lyon (Lyon, France).
This study was supported by grants from Agence Nationale pour la Recherche centre le SIDA et les Hepatites Virales (ANRS), Agence Nationale de la Recherche (ANR; Physio-WAS-ANR-07-MRAR-022-03), Agence Française centre la myopathie (AFM), and the European Community (FP7-HEALTH-2007-B/222878 “PERSIST,” E-RARE-06-01 “GETHERTHAL,” ERC-2008-AdG-233130-HEPCENT). C.F. was supported by an ANRS postdoctoral fellowship. J.A.B and P.R. are supported by FP7-HEALTH-2007-B/222878 “PERSIST,” Programa de Fomento de Cooperación Científica Internacional (110-90.1), Plan Nacional de Salud y Farmacia (SAF 2009-07 164), Instituto de Salud Carlos III (RETICS-RD06/0010/0015), and Fundación Botín.
Authorship
Contribution: E.V. coordinated the project, designed and performed the experiments, analyzed the data, and wrote the manuscript; F.-L.C. designed the experiments, analyzed the data, and wrote the manuscript; C.F., C.C., D.N., F.A., and P.R. performed the experiments; and D.T. and J.B. provided hBM samples, technical advice, and help with the mice.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Els Verhoeyen and F. L. Cosset, EVIR, Inserm U758, ENS de Lyon, 46 Allée d'Italie, 69364 Lyon Cedex 07, France; e-mail: els.verhoeyen@ens-lyon.fr, flcosset@ens-lyon.fr.
References
Author notes
C.F. and C.C. contributed equally to this work.
F.-L.C. and E.V. contributed equally to this work and are co–corresponding authors.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal