Abstract
CD4+ Th cells are pivotal for the generation and maintenance of CD8+ T-cell responses. “Helped” CD8+ T cells receive signals during priming that prevent the induction of the proapoptotic molecule TNF-related apoptosis-inducing ligand (TRAIL) during reactivation, thereby enabling robust secondary expansion. Conversely, “helpless” CD8+ T cells primed in the absence of Th induce TRAIL expression after restimulation and undergo activation-induced cell death. In the present study, we investigated the molecular basis for the differential regulation of TRAIL in helped versus helpless CD8+ T cells by comparing their transcriptional profiles, and have identified a transcriptional corepressor, NGFI-A binding protein 2 (Nab2), that is selectively induced in helped CD8+ T cells. Enforced expression of Nab2 prevents TRAIL induction after restimulation of primary helpless CD8+ T cells, and expression of a dominant-negative form of Nab2 in helped CD8+ T cells impairs their secondary proliferative response that is reversible by TRAIL blockade. Finally, we observe that the CD8+ T-cell autocrine growth factor IL-2 coordinately increases Nab2 expression and decreases TRAIL expression. These findings identify Nab2 as a mediator of Th-dependent CD8+ T-cell memory responses through the regulation of TRAIL and the promotion of secondary expansion, and suggest a mechanism through which this operates.
Introduction
Cytotoxic CD8+ T cells play a fundamental role in the defense against viral and intracellular bacterial infections through the generation of early effectors that eradicate infected cells and of long-lived memory cells that confer durable protection against recurring infection.1-3 CD4+ Th cells influence the generation and maintenance of CD8+ T-cell responses at several levels. This includes the recruitment of naive cells to dendritic cells within lymph nodes during priming, insuring their survival after primary expansion and facilitating the migration of effector cells into peripheral sites of Ag re-encounter.4-7 In addition to these functions, Th cells are instrumental in the generation of CD8+ T-cell memory through activation of APCs via CD40L-CD40 interactions to a state in which they can prime CD8+ T cells capable of secondary expansion after Ag reencounter (reviewed by Bevan8 ). Several studies have demonstrated that this process involves modification of the differentiation program in CD8+ T cells by signals received during the initial Ag encounter, resulting in specific patterns of gene expression in their daughter cells.9-11 The clonal progeny of helpless CD8+ T cells, for example, induce the proapoptotic molecule TNF-related apoptosis-inducing ligand (TRAIL) and its receptor (DR5) after restimulation, and subsequently undergo activation-induced cell death (AICD).12-14 Helped CD8+ T cells, in contrast, do not induce TRAIL expression after restimulation, and instead undergo the robust secondary T-cell responses associated with immune memory.13
In the present study, we investigated the molecular mechanisms regulating TRAIL expression in CD8+ T cells by comparing the transcriptional profile of reactivated helped versus helpless CD8+ T cells. This led to the identification of a transcriptional corepressor of TRAIL expression, NGFI-binding protein 2 (Nab2),15 which is selectively induced in helped, but not helpless, CD8+ T cells after restimulation. Exogenous expression of Nab2 effectively suppressed the induction of TRAIL in restimulated helpless CD8+ T cells, and inhibition of Nab2 function in helped CD8+ T cells prevented their ability to undergo secondary expansion, and this could be restored by blockade of TRAIL. Lastly, we found that the addition of IL-2, a key autocrine factor that can rescue the secondary response defect in helpless CD8+ T cells, can induce Nab2 expression and prevent TRAIL. These data identify Nab2 as a molecular mediator of Th-dependent CD8+ T-cell memory through regulation of TRAIL expression.
Methods
Mice and cell culture
C57BL/6J mice were purchased from The Jackson Laboratory. TCR-transgenic OT-I mice and C57BL/6J recipient mice were bred in-house. All animal experiments were performed in accordance with institutional and national guidelines of all participating institutions.
All cells were cultured in IMDM (GIBCO-BRL) supplemented with 8% FCS, 50μM 2-mercaptoethanol, 2mM l-glutamine, 20 U/mL of penicillin, and 20 μg/mL of streptomycin.
Generation of in vivo primed polyclonal CD8+ T cells
Helped and helpless E1B192-200–specific CD8+ T cells were generated as described previously.16 Briefly, C57BL/6J mice were treated with 100 μg of GK1.5 administered intraperitoneally (helpless) or were left untreated (helped) before subcutaneous immunization with 1 × 107 irradiated (3000 rad) TAP−/−Ad5E1-MEC. Three days after immunization, all mice were treated with 100 μg of GK1.5. Spleens and draining lymph nodes were harvested 7 days after immunization, and CD8+ T cells were purified with the CD8 negative isolation kit (Miltenyi Biotec) according to the manufacturer's protocol. Purity was 88%-95%.
Microarray and data analysis
Purified CD8+ T cells from helped and helpless mice were sorted on CD44hi expression by flow cytometry. Cells were restimulated for 4 hours with 2 μg/mL of E1B192-200 peptide in the presence of 20μM qVD-OPh (R&D Systems). RNA was extracted using TRIzol reagent (Gibco BRL) according to the manufacturer's instructions. Five micrograms of total RNA was used to generate cDNA with RT Superscript III (Invitrogen). cDNA was labeled indirectly using cyanine 3 and cyanine 5 (Amersham). Labeled samples were purified and spotted in an equimolar ratio on the Mouse Exonic Evidence Based Oligonucleotide (MEEBO) array. Images were preview scanned on an Axon 4000B and channels were balanced until an overall Cy5/Cy3 ratio of 1 was achieved before a final data scan at 5 μm was performed in GenePix. The images were manually gridded, and then saved for analysis. Data were analyzed using the Spotfire Decision Site.
Gene Ontology terms were downloaded from BioMart (www.biomart.org, Version NCBI37, April 9, 2009). Genes with Biologic Process term “regulation of transcription” (GO:0045449) were assumed to encode transcription factors. Of 276 probes with an absolute log2 ratio > 2, 231 probes were present in BioMart; the remaining cDNA probes did not match currently known genes. All microarray data are available at the Gene Expression Omnibus (GEO) under accession number GSE33862.
Real-time RT-PCR
Purified CD8+ T cells were restimulated with 2 μg/mL of E1B192-200 peptide for 4 hours or were left untreated. Where indicated, 20 CU/mL of human recombinant IL-2 was added during peptide restimulation. Cells were washed twice with PBS before total RNA was isolated using TRIzol reagent according to the manufacturer's protocol. RNA was reverse transcribed with RT SuperScript II (Invitrogen) using random hexamers (GIBCO-BRL) in the presence of RNAseOUT (Invitrogen). Sequence-specific primers for murine IFNγ, TRAIL, Nab2, L32, and 18S were described previously.17 Real-time RT-PCR was performed in a Bio-Rad iQ5 thermal cycler using the SyBr Green detection protocol as outlined by the manufacturer (Abgene). L32 and 18S were used as internal controls.
Plasmids, transfections, and retroviral transductions
Murine Nab2 cDNA (BC045139; Open Biosystems) was cloned into pcDNA3.1 to generate pcDNA3.1-Nab2. Nab2E51K was generated with site-directed mutagenesis according to the manufacturer's protocol (Promega) and cloned into pMIG to generate pMIG-Nab2E51K. Sequences were confirmed by sequence analysis.
For exogenous expression of Nab2 in helpless CD8+ T cells, 10 × 106 cells were transfected with 2 μg of pcDNA3.1-Nab2 or the empty pcDNA3.1 control, together with 0.5 μg of pmaxGFP according to the manufacturer's protocol (Amaxa). At 24 hours after transfection, cells were harvested and restimulated with 2 μg/mL of E1B192-200 peptide for 4 hours or were left untreated before RNA isolation.
The production of retrovirus containing pMIG-NAB2E51K or pMIG-EV for the transduction of OT-I thymocytes was performed as described previously.18
T-cell transfer of naive NAB2E51K-expressing OTI-T cells
OT-I Ly5.1 thymocytes were retrovirally transduced with pMIG-NAB2E51K, or pMIG-EV, as described previously.18 Twenty-four hours after retroviral transduction, 0.5-1 × 106 green fluorescent protein (GFP)–sorted OT-I thymocytes were transferred intrathymically into C57BL/6 carrier mice. Two weeks after transfer, spleens and lymph nodes were collected and CD8+ T cells were purified (Mouse CD8 T Lymphocyte Enrichment Set; BD Biosciences). Approximately 2 × 106 CD8+ T cells containing 60 GFP+ OT-I cells were transferred into C57BL/6J mice. The fraction of transferred GFP-expressing OT-I cells within the isolated CD8+ T cells was confirmed by flow cytometry. The next day, experimental mice received 100 μg of GK1.5 IP or were left untreated, and 3 days later, they were immunized with 2 × 106 irradiated (1500 rad) OVA-coated splenocytes as described previously.19 All mice were treated with GK1.5 3 days after immunization. Spleens and lymph nodes were harvested 7 days after immunization, and the number of OVA257-264–specific CD8+ T cells (GFP+ and GFP−) was determined by Kb-OVA257-264 tetramer staining. To assess the fold expansion of OVA257-264–specific CD8+ T cells, the absolute number of Kb-OVA257-264 tetramer+ CD8+ T cells obtained after 7 days in vitro culture with irradiated (3000 rad) MEC.B7.SigOVA cells in a ratio of 40:1 was divided by the absolute number of Kb-OVA257-264 tetramer+ CD8+ T cells placed into culture, as described previously.16 For blocking studies, CD8+ T cells were sorted by CD8 negative selection (Miltenyi Biotec) before in vitro restimulation with MEC.B7.SigOVA cells in a effector:stimulator ratio of 25:1 in the presence of 5 μg/mL of murine TRAIL-R-FC or Fas-FC (R&D Systems) or 20 CU of recombinant human IL-2.
Flow cytometry and analysis
mAbs against CD8, CD44, and IFNγ (BD Pharmingen and eBioscience) were used for flow cytometry. Kb-OVA257-264 tetramers were generated as described previously.20 For intracellular cytokine staining, 1-1.5 × 106 cells were cultured in the absence or presence of 2 μg/mL E1B192-200 peptide or 1 μg/mL of OVA257-264 and brefeldin A for 5 hours at 37°C. After surface Ab staining, cells were fixed and permeabilized for intracellular IFNγ staining according to the manufacturer's protocol (BD Pharmingen). Flow cytometry samples were acquired on a FACSCalibur instrument (BD Biosciences) and analyzed using FlowJo Version 8 software (TreeStar).
Statistical analysis
Results
Differential expression of Nab2 in helped versus helpless CD8+ T cells
To elucidate how TRAIL expression is regulated in helped versus helpless CD8+ T cells, we first sought to identify the molecular regulators that are differentially expressed on secondary antigenic challenge. To this end, Ag-specific CD8+ T cells were generated using an established cross-priming model (TAP−/−Ad5E1-MEC) that has been shown previously to induce a CD4-dependent secondary CD8+ T-cell response specific for the immunodominant epitope (E1B192-200).16,21 Eight days after immunization of CD4-depleted (helpless) or intact (helped) mice, spleens and lymph nodes were harvested and CD8+ T cells were isolated by positive selection. Ag-experienced CD8+ T cells were further enriched for CD44hi expression by flow cytometry. As assessed by intracellular IFNγ staining after antigenic restimulation, we estimated an enrichment of E1B192-200–specific CD8+ T cells of 30- to 50-fold with this isolation method (Figure 1A and data not shown). Using a selection method that did not involve TCR engagement, reactivation of the CD8+ T cells during isolation was minimized.
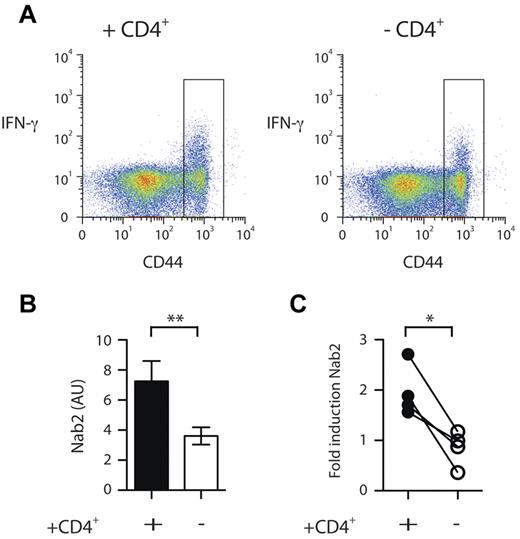
Nab2 is differentially regulated in helped versus helpless CD8+ T cells. Spleens from TAP−/−Ad5E1-MEC–primed mice in a helped or helpless setting were harvested and CD8+ T cells isolated. A CD44hi sort was performed on nonreactivated cells. (A) Purified CD8+ T cells were restimulated for 4 hours with the E1B192-200 peptide and intracellular IFNγ staining was performed. Nineteen percent and 10% IFNγ+ cells for helped and helpless CD44hi CD8+ T cells was measured, respectively. Gate represents sorting gate employed to enrich for CD44hi CD8+ T cells. (B) Quantitative RT-PCR analysis of Nab2 mRNA levels in restimulated CD44hi-sorted CD8+ T cells. (C) Helped and helpless CD8+ T cells isolated from TAP−/− Ad5E1-MEC–primed mice were restimulated with the E1B192-200 peptide or left untreated, and the fold induction of Nab2 transcripts was determined. Each connected dataset represents an independently performed experiment (n = 4; T cells were pooled from 4 mice per group). *P < .05; **P < .02.
Nab2 is differentially regulated in helped versus helpless CD8+ T cells. Spleens from TAP−/−Ad5E1-MEC–primed mice in a helped or helpless setting were harvested and CD8+ T cells isolated. A CD44hi sort was performed on nonreactivated cells. (A) Purified CD8+ T cells were restimulated for 4 hours with the E1B192-200 peptide and intracellular IFNγ staining was performed. Nineteen percent and 10% IFNγ+ cells for helped and helpless CD44hi CD8+ T cells was measured, respectively. Gate represents sorting gate employed to enrich for CD44hi CD8+ T cells. (B) Quantitative RT-PCR analysis of Nab2 mRNA levels in restimulated CD44hi-sorted CD8+ T cells. (C) Helped and helpless CD8+ T cells isolated from TAP−/− Ad5E1-MEC–primed mice were restimulated with the E1B192-200 peptide or left untreated, and the fold induction of Nab2 transcripts was determined. Each connected dataset represents an independently performed experiment (n = 4; T cells were pooled from 4 mice per group). *P < .05; **P < .02.
The enriched helped and helpless CD8+ T cells were then restimulated for 4 hours in vitro with the E1B192-200 peptide, and their transcriptional profile was analyzed with the Mouse Exonic Evidence Based Oligonucleotide (MEEBO) array platform.22 Comparison of these transcriptional profiles revealed that 276 of 30 000 probes displayed a differential expression pattern of at least a 2-fold difference in helpless CD8+ T cells compared with helped CD8+ T cells (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Because our interest was in gene regulation of TRAIL, we focused on the 36 genes that were assigned to the category “regulation of transcription” by the BioMart gene ontology list.23 Of these, 20 genes were up-regulated and 16 were down-regulated in helpless CD8+ T cells compared with helped CD8+ T cells (Table 1). We further examined transcriptional regulators implicated in cell survival. An intriguing candidate in this category was Nab2 (Table 1), because its homologous family member Nab1 has been shown to regulate several apoptosis genes, including TRAIL, in epithelial cells through interaction with the early growth response genes.17 Using a semiquantitative RT-PCR approach, we confirmed that the expression of Nab2 was significantly reduced in reactivated helpless versus helped CD8+ T cells (Figure 1B; P = .005). Furthermore, we found that Nab2 transcripts were increased by approximately 1.5- to 3-fold after antigenic restimulation of helped CD8+ T cells. However, because we studied a polyclonal CD8+ T-cell response that contained a mixture of Ag-specific and non–Ag-specific CD8+ T cells, this number may be an underestimate. Helpless CD8+ T cells, in contrast, failed to show induction of Nab2 expression (Figure 1C; P = .02). These data reveal that the presence of CD4+ T-cell help during the priming of CD8+ T cells positively modifies the expression pattern of Nab2 in their clonal progeny.
Differential expression of transcription-regulating genes in reactivated helpless versus helped CD8+ T cells
| Reference no. . | Gene . | Log2 ratio . | Reference no. . | Gene . | Log2 ratio . |
|---|---|---|---|---|---|
| NM_133208 | Zfp287 | −1.6074 | NM_008950 | Psmc5 | 1.0019 |
| NM_145546 | Gtf2b | −1.4275 | NM_183298 | Foxe1 | 1.0262 |
| NM_011445 | Sox6 | −1.3916 | NM_008391 | Irf2 | 1.0305 |
| NM_177338 | Hmbox1 | −1.3042 | NM_009558 | Zfp51 | 1.0320 |
| NM_008668 | Nab2 | −1.2733 | NM_010136 | Eomes | 1.0570 |
| NM_172716 | Pcgf3 | −1.2002 | NM_177888 | Zfp78 | 1.0821 |
| NM_008651 | Mybl1 | −1.1843 | NM_177888 | Zfp78 | 1.0821 |
| NM_009986 | Cutl1 | −1.1471 | NM_023434 | 5730589K01Rik | 1.0865 |
| NM_010432 | Hipk1 | −1.1431 | NM_153287 | Axud1 | 1.1372 |
| NM_175477 | Zfp574 | −1.1397 | NM_030676 | Nr5a2 | 1.1422 |
| NM_010731 | Zbtb7a | −1.0740 | NM_172495 | Ncoa7 | 1.1723 |
| NM_023266 | Zfp120 | −1.0630 | NM_011050 | Pdcd4 | 1.1815 |
| NM_175247 | Zfp28 | −1.0398 | NM_133966 | Taf5l | 1.1948 |
| NM_011768 | Zfx | −1.0305 | NM_145455 | Btf3 | 1.2371 |
| NM_027063 | 1700013G24Rik | −1.0208 | NM_010905 | Nfia | 1.3377 |
| NM_201355 | BC047219 | −1.0031 | NM_010411 | Hdac3 | 1.3448 |
| NM_033552 | Slc4a10 | 1.4431 | |||
| NM_017376 | Tef | 1.5242 | |||
| NM_183185 | D930016N04Rik | 1.6470 | |||
| NM_019935 | Ovol1 | 1.7046 | |||
| NM_009372 | Tgif | 1.9097 |
| Reference no. . | Gene . | Log2 ratio . | Reference no. . | Gene . | Log2 ratio . |
|---|---|---|---|---|---|
| NM_133208 | Zfp287 | −1.6074 | NM_008950 | Psmc5 | 1.0019 |
| NM_145546 | Gtf2b | −1.4275 | NM_183298 | Foxe1 | 1.0262 |
| NM_011445 | Sox6 | −1.3916 | NM_008391 | Irf2 | 1.0305 |
| NM_177338 | Hmbox1 | −1.3042 | NM_009558 | Zfp51 | 1.0320 |
| NM_008668 | Nab2 | −1.2733 | NM_010136 | Eomes | 1.0570 |
| NM_172716 | Pcgf3 | −1.2002 | NM_177888 | Zfp78 | 1.0821 |
| NM_008651 | Mybl1 | −1.1843 | NM_177888 | Zfp78 | 1.0821 |
| NM_009986 | Cutl1 | −1.1471 | NM_023434 | 5730589K01Rik | 1.0865 |
| NM_010432 | Hipk1 | −1.1431 | NM_153287 | Axud1 | 1.1372 |
| NM_175477 | Zfp574 | −1.1397 | NM_030676 | Nr5a2 | 1.1422 |
| NM_010731 | Zbtb7a | −1.0740 | NM_172495 | Ncoa7 | 1.1723 |
| NM_023266 | Zfp120 | −1.0630 | NM_011050 | Pdcd4 | 1.1815 |
| NM_175247 | Zfp28 | −1.0398 | NM_133966 | Taf5l | 1.1948 |
| NM_011768 | Zfx | −1.0305 | NM_145455 | Btf3 | 1.2371 |
| NM_027063 | 1700013G24Rik | −1.0208 | NM_010905 | Nfia | 1.3377 |
| NM_201355 | BC047219 | −1.0031 | NM_010411 | Hdac3 | 1.3448 |
| NM_033552 | Slc4a10 | 1.4431 | |||
| NM_017376 | Tef | 1.5242 | |||
| NM_183185 | D930016N04Rik | 1.6470 | |||
| NM_019935 | Ovol1 | 1.7046 | |||
| NM_009372 | Tgif | 1.9097 |
Probes displaying an absolute log2 ratio of > 1 were analyzed for genes with the annotation “regulation of transcription” (GO:0045449) by BioMart; Log2 ratio > −1, lower expression in helpless CD8+ T cells; log2 ratio > 1, higher expression in helpless CD8+ T cells.
Nab2 blocks TRAIL expression in reactivated helpless CD8+ T cells
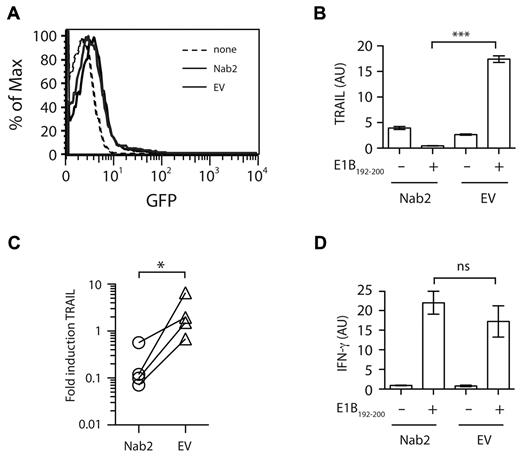
Having correlated Nab2 with the absence of TRAIL expression in restimulated helped CD8+ T cells, we next assessed whether forced expression of Nab2 could similarly regulate TRAIL induction in helpless cells. Freshly isolated helpless CD8+ T cells from TAP−/−Ad5E1-MEC–immunized mice were transfected with an expression plasmid encoding Nab2 or control empty vector (EV), and assessed 24 hours later for TRAIL expression after reactivation with the cognate Ag (Figure 2). Strikingly, exogenous expression of Nab2 abolished the induction of TRAIL transcripts in helpless CD8+ T cells after Ag encounter, in contrast to the marked induction of TRAIL mRNA detected in CD8+ T cells transfected with the EV (Figure 2B-C, P = .0002 and P = .029, respectively). Nab2 expression did not appear to affect the overall functional response to antigenic stimuli, because both Nab2- and EV-transfected CD8+ T cells displayed comparable levels of IFNγ mRNA induction after secondary Ag encounter (Figure 2D). These results show that Nab2 can prevent the TRAIL induction that occurs after restimulation of helpless CD8+ T cells.
Nab2 blocks TRAIL induction in restimulated helpless CD8+ T cells. CD8+ T cells were purified from TAP−/−Ad5E1-MEC–primed, helpless mice and were transfected with 2 μg of pcDNA3.1-Nab2 or the empty control vector, together with 0.5 μg of the reporter plasmid pMAX-GFP. (A) Twenty-four hours after transfection, GFP expression was determined as a measurement for transfection efficiency. (B-D) CD8+ T cells were cultured for 4 hours in the presence or absence of the E1B192-200 peptide, and the relative expression of TRAIL (B) and IFNγ (D) mRNA was determined. (C) The fold induction of TRAIL in Nab2- or EV-transfected CD8+ T cells was assessed by comparing mRNA levels before and after E1B192-200 peptide restimulation. Each connected dataset represents an independently performed experiment (n = 4; T cells were pooled from 4 mice per group). *P < .05; ***P < .005.
Nab2 blocks TRAIL induction in restimulated helpless CD8+ T cells. CD8+ T cells were purified from TAP−/−Ad5E1-MEC–primed, helpless mice and were transfected with 2 μg of pcDNA3.1-Nab2 or the empty control vector, together with 0.5 μg of the reporter plasmid pMAX-GFP. (A) Twenty-four hours after transfection, GFP expression was determined as a measurement for transfection efficiency. (B-D) CD8+ T cells were cultured for 4 hours in the presence or absence of the E1B192-200 peptide, and the relative expression of TRAIL (B) and IFNγ (D) mRNA was determined. (C) The fold induction of TRAIL in Nab2- or EV-transfected CD8+ T cells was assessed by comparing mRNA levels before and after E1B192-200 peptide restimulation. Each connected dataset represents an independently performed experiment (n = 4; T cells were pooled from 4 mice per group). *P < .05; ***P < .005.
A dominant-negative Nab2 (Nab2E51K) prevents the secondary expansion of helped CD8+ T cells via TRAIL regulation
Because TRAIL deficiency rescues reactivated helpless CD8+ T cells from AICD,12-14 and because Nab2 potently suppresses their induction of TRAIL (Figure 2B-C), we hypothesized that inhibition of endogenous Nab2 activity might interfere with the secondary proliferation response of helped CD8+ T cells. To address this, we introduced a dominant-negative form of Nab2 (Nab2E51K) into OT-I transgenic CD8+ T cells together with GFP on a bicistronic vector. Nab2E51K retains the ability to oligomerize with endogenous Nab proteins, but is incapable of interacting with, and thereby repressing, its binding partners, the Egr transactivators.24 Because Th-cell–dependent programming of memory functions—including secondary expansion—occurs during the first days of CD8+ T-cell priming, it was necessary that the Nab2E51K-expressing CD8+ T cells be naive before their initial Ag encounter.9,16,25 We therefore generated naive Nab2E51K- or EV-expressing primary OT-I T cells by retroviral transduction into OT-I thymocytes, followed by intrathymic transfer into new donor mice to allow for the completion of T-cell differentiation.18 The peripheral OT-I/Nab2E51K cells and OT-I/EV control cells displayed a naive T-cell phenotype, as assessed by their low-intermediate expression levels of CD44 and high expression levels of CD62L when harvested 2 weeks later from the spleens and lymph nodes of recipient mice (supplemental Figure 1).
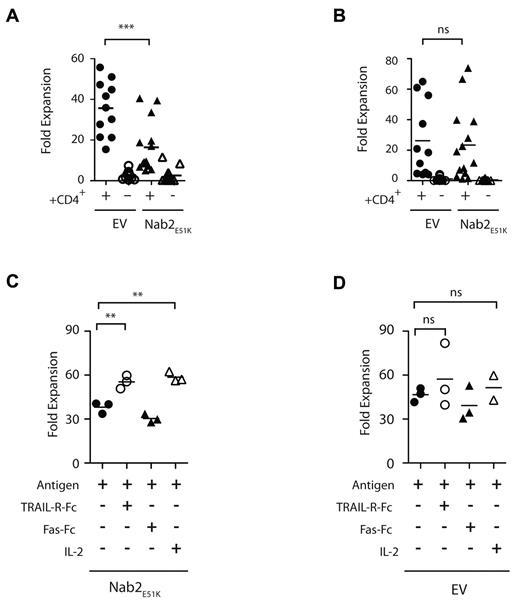
Approximately 60 GFP+, OT-I/Nab2E51K or OT-I/EV T cells were transferred into naive recipients to obtain OT-I T-cell responses phenotypically comparable to the endogenous polyclonal repertoire.26,27 Mice were immunized with OVA-coated splenocytes as described previously19 in a helped or helpless setting, and 7 days later, the OVA257-264–specific T-cell responses in the spleens and lymph nodes were enumerated ex vivo. A portion of the spleen and lymph node populations were restimulated in vitro with MEC.B7.SigOVA cells and re-evaluated for OVA257-264–specific T cells, with the secondary proliferative response calculated by comparing the input and output number of OVA257-264–specific T cells.16 Consistent with our finding that Nab2 represses the expression of TRAIL (Figure 2), blocking Nab2 function in helped OT-I T cells markedly reduced their secondary proliferative response (Figure 3A; P = .001). This defect was cell intrinsic for Nab2E51K-expressing OT-I T cells, because both the OT-I/EV T cells and the endogenous OVA257-264–specific CD8+ T cells proliferated substantially after secondary Ag encounter in helped mice (Figure 3A-B). These data reveal that helped CD8+ T cells require Nab2 function to mount an optimal secondary T-cell expansion.
Nab2E51K affects CD8+ T-cell expansion after restimulation mediated through TRAIL. Sixty naive OTI T cells expressing Nab2E51K or the EV were transferred into C57BL/6J mice before immunization with OVA-loaded splenocytes in a helped or helpless setting. Seven days later, lymphocytes were harvested and the number of OVA257-264–specific T cells was determined by Kb-OVA257-264 tetramer staining. Cells were restimulated with MEC.B7.SigOVA cells for 7 days, and the number of OVA257-264–specific T cells was reassessed. The -fold expansion was determined for (A) exogenous Nab2E51K-expressing or EV-expressing OT-I T cells (A) and for the corresponding endogenous OVA257-264-specific CD8+ T-cell responses (B). Each data point represents an individual mouse or 2 pooled mice (n = 3). Graph shows data compiled from 3 individual experiments (GFP WT, n = 11; all other groups, n = 14). (C-D) CD8+ T cells were isolated from helped and helpless mice that had received OT-I-Nab2E51K cells (C) or OT-I-EV cells (D) before immunization with OVA-coated splenocytes. CD8+ T cells were restimulated with MEC.B7.SigOVA cells for 7 days in the presence of 5 μg/mL of TRAL-FC, 5 μg/mL of Fas-FC, or 20 CU/mL of recombinant human IL-2 or medium alone (ctrl). The secondary proliferation of GFP+ OTI T cells was assessed with Kb-OVA257-264 tetramer staining, as described previously.23 Secondary expansion of helped OT1-EV cells with Ag alone was significantly higher than that of Nab2E51K–expressing T cells (P < .05; supplemental Figure 2). Each data point represents T cells pooled from 2 mice; the figure depicts 3 biologic replicates. **P < .02; ***P < .002.
Nab2E51K affects CD8+ T-cell expansion after restimulation mediated through TRAIL. Sixty naive OTI T cells expressing Nab2E51K or the EV were transferred into C57BL/6J mice before immunization with OVA-loaded splenocytes in a helped or helpless setting. Seven days later, lymphocytes were harvested and the number of OVA257-264–specific T cells was determined by Kb-OVA257-264 tetramer staining. Cells were restimulated with MEC.B7.SigOVA cells for 7 days, and the number of OVA257-264–specific T cells was reassessed. The -fold expansion was determined for (A) exogenous Nab2E51K-expressing or EV-expressing OT-I T cells (A) and for the corresponding endogenous OVA257-264-specific CD8+ T-cell responses (B). Each data point represents an individual mouse or 2 pooled mice (n = 3). Graph shows data compiled from 3 individual experiments (GFP WT, n = 11; all other groups, n = 14). (C-D) CD8+ T cells were isolated from helped and helpless mice that had received OT-I-Nab2E51K cells (C) or OT-I-EV cells (D) before immunization with OVA-coated splenocytes. CD8+ T cells were restimulated with MEC.B7.SigOVA cells for 7 days in the presence of 5 μg/mL of TRAL-FC, 5 μg/mL of Fas-FC, or 20 CU/mL of recombinant human IL-2 or medium alone (ctrl). The secondary proliferation of GFP+ OTI T cells was assessed with Kb-OVA257-264 tetramer staining, as described previously.23 Secondary expansion of helped OT1-EV cells with Ag alone was significantly higher than that of Nab2E51K–expressing T cells (P < .05; supplemental Figure 2). Each data point represents T cells pooled from 2 mice; the figure depicts 3 biologic replicates. **P < .02; ***P < .002.
We also sought to determine whether the mechanism through which Nab2E51K blocks secondary expansion in helped OT-I T cells involved TRAIL. To accomplish this, OT-I/Nab2E51K T cells were restimulated in the presence of a soluble form of the TRAIL receptor TRAIL-R-Fc, as described previously.13,28 To preclude indirect effects of blocking TRAIL, CD8+ T cells were purified before in vitro restimulation. After this treatment, the secondary expansion of OT-I/Nab2E51K cells was significantly reduced compared with OT1-EV cells (supplemental Figure 2; P < .05). Strikingly, when TRAIL activity was blocked during restimulation, the expansive capacity of OT-I/Nab2E51K T cells was restored (Figure 3C; P < .02), supporting our hypothesis that blocking Nab2 in helped CD8+ T cells results in reduced secondary expansion because of TRAIL-mediated AICD. The effect of blocking TRAIL was similar to the rescue observed after IL-2 addition, which was shown previously to rescue secondary expansion of helpless CD8+ T cells in part by interfering with TRAIL-mediated AICD13,28 (Figure 3C; P < .02). A control fusion protein, Fas-Fc, failed to restore secondary responses (Figure 3C), and the addition of TRAIL-R-FC did not significantly alter the expansion of OT-I/EV control–expressing T cells compared with the expansion in the presence of Ag alone (Figure 3D controls; P = .47). These data reveal that in the absence of Nab2 function, helped CD8+ T cells undergo TRAIL-mediated AICD in a manner similar to helpless CD8+ T cells that fail to induce Nab2.
IL-2 induces Nab2 expression in helpless CD8+ T cells
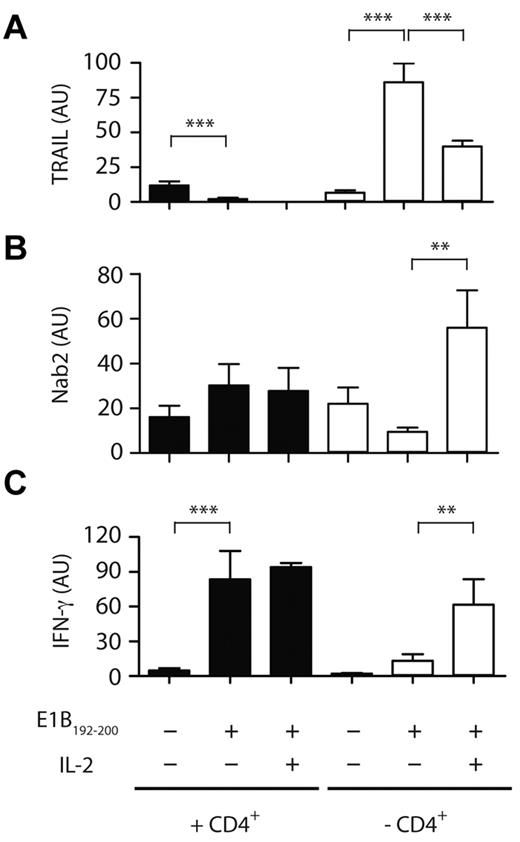
We next investigated the factors that may govern the induction of Nab2 in reactivated CD8+ T cells. Previous studies have demonstrated that CD8+ T cells must receive IL-2 signals during priming to allow for secondary expansion after Ag re-encounter, and we and others have recently shown that CD8+ T cells themselves are the source of this required cytokine.29,30 In addition, IL-2 signaling restores the proliferative and functional capacity of helpless CD8+ T cells after reactivation, and this occurs by inhibition of TRAIL expression.13,28 We therefore speculated that the mechanism through which IL-2 prevents TRAIL-mediated AICD is through the induction of Nab2 expression. To address this, we investigated whether the addition of exogenous IL-2 at restimulation would prevent TRAIL induction in helpless E1B192-200–specific CD8+ T cells. In the experiment shown in Figure 4A, helped CD8+ T cells expressed low levels of TRAIL message ex vivo, and antigenic stimulation further suppressed TRAIL mRNA levels (P < .005). In contrast, helpless CD8+ T cells rapidly up-regulated TRAIL message in response to Ag (P < .0001), which was inhibited by approximately 50% after the addition of 20 ng/mL of IL-2 during antigenic restimulation (P < .005). We then assessed whether IL-2 affects the expression of TRAIL via Nab2. Exogenous IL-2 increased the level of Nab2 transcripts in restimulated helpless CD8+ T cells by approximately 4-fold (Figure 4B; P < .01). The addition of IL-2 was accompanied by an increase in IFNγ transcripts in helpless CD8+ T cells (Figure 4C), but did not increase Nab2 or IFN-γ transcription above the high basal levels found in restimulated helped CD8+ T cells, suggesting that secondary Ag encounter sufficed for helped CD8+ T cells to induce proficient effector T cells (Figure 4B-C). These data establish a link between IL-2 signaling and TRAIL expression in CD8+ T cells that is mediated by Nab2.
IL-2 affects the transcriptional profile of TRAIL and Nab2 in helpless CD8+ T cells. Purified CD8+ T cells isolated from helped (filled bars) or helpless mice (open bars) were restimulated with the E1B192-200 peptide in the absence or presence of 20 CU/mL of recombinant human IL-2 or were left untreated. The relative mRNA expression of TRAIL (A), Nab2 (B), and IFNγ (C) was determined by RT-PCR (T cells were pooled from 3-4 mice per group and data shown are representative of 3 experiments). **P < .02; ***P < .005.
IL-2 affects the transcriptional profile of TRAIL and Nab2 in helpless CD8+ T cells. Purified CD8+ T cells isolated from helped (filled bars) or helpless mice (open bars) were restimulated with the E1B192-200 peptide in the absence or presence of 20 CU/mL of recombinant human IL-2 or were left untreated. The relative mRNA expression of TRAIL (A), Nab2 (B), and IFNγ (C) was determined by RT-PCR (T cells were pooled from 3-4 mice per group and data shown are representative of 3 experiments). **P < .02; ***P < .005.
Discussion
The results of the present study demonstrate that the transcriptional regulator Nab2 suppresses TRAIL expression in reactivated CD8+ T cells, thereby allowing secondary proliferative responses to occur. Helpless CD8+ T cells made to express Nab2 at secondary antigenic stimulation were able to avoid TRAIL induction. In addition, blockade of Nab2 function in helped CD8+ T cells prevented the secondary proliferation that is a hallmark of the memory response, and instead directed them toward TRAIL-mediated AICD, as did their helpless counterparts. Lastly, we found that IL-2 induces Nab2 and prevents TRAIL induction in restimulated helpless CD8+ T cells, thereby providing a mechanism for the recent findings on IL-2–mediated rescue of the helpless phenotype.13,28 These results provide the first insights into the molecular components through which Th enables secondary expansion in CD8+ T cells through regulation of TRAIL.
Previous studies examining the transcriptional regulation of helpless CD8+ T cells identified T-bet as an attenuator of CD4+ T-cell help during T-cell priming, thereby negatively affecting the effective memory T-cell development.31 Furthermore, in vitro T-cell activation studies have shown that lack of CD4+ T-cell help results in a diminished histone acetylation of the IFNγ promoter that prevents the development of functional responses of helpless CD8+ T cells.32,33 In the present study, we describe Nab2 as a transcriptional regulator that manifests the effects of CD4+ T-cell help for CD8+ T cells, allowing the development of secondary CD8+ T-cell responses to occur. In support of this notion, we found that Nab2 can be induced by IL-2, which itself is required as an autocrine factor for CD8+ T-cell memory.
A variety of cognate and cytokine signals have been proposed to be involved in communicating the “help message” during T-cell priming, including CD40-CD40L, CD28-CD80/86, CD27-CD70, and IL-2R signaling; however, which of these signals synergizes with TCR engagement to program Nab2 induction on recall is to date unclear.12,29,34-39 A possible link with IL-2 is compelling, given our recent finding that autocrine IL-2 is critical for secondary expansion in helped CD8+ T cells, and the present study's demonstration that IL-2 can induce Nab2 and inhibit TRAIL in helpless CD8+ T cells (Figure 4B). Interestingly, Nab2 can regulate the production of IL-2 in activated T cells, and both Nab2 and the transcription factor Egr-1 have been shown to interact with the IL-2 promoter in activated T cells.40,41 These data suggest that Nab2 and IL-2 are components of a positive feedback loop through which Th signals provided during priming are integrated into the memory response of CD8+ T cells through regulation of TRAIL-mediated AICD.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the staff of the flow cytometry facilities of the St Jude Children's Research Hospital and the National Cancer Institute for cell sorting, the microarray facility of St Jude Children's Research Hospital for performing the array, Dr M. Wang (St Jude Children's Research Hospital) and Dr B. van Steensel (National Cancer Institute) for assisting with the analysis of microarray data, and Dr N. Droin for the RT-PCR primers.
This study was supported by grants from the National Institutes of Health (RO1 AI076972 and RO1CA81261 to S.P.S.), the Leukemia & Lymphoma Society (1630-06 to S.P.S.), the Cancer Research Institute/Irvington Fellowship, and the Dutch Science Foundation (VENI grant 916.76.127 to M.C.W.).
National Institutes of Health
Authorship
Contribution: M.C.W., C.G., E.M.J., D.R.G., T.N.S., J.P.M., and S.P.S. designed the experiments; M.C.W., C.G., R.A., E.M.J., and P.F. performed the experiments and analyzed the data; and M.C.W., D.R.G., and S.P.S. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for M.C.W. is Department of Hematopoiesis, Sanquin Research/Landsteiner Laboratory AMC, Amsterdam, The Netherlands.
Correspondence: M. Wolkers, Department of Hematopoiesis, Sanquin Research/Landsteiner Laboratory AMC, Plesmanlaan 125, 1066CX Amsterdam, The Netherlands; e-mail: m.wolkers@sanquin.nl.
References
Author notes
D.R.G. and S.P.S. share senior authorship.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal