Abstract
Osteopontin (OPN) is a glycoprotein that is secreted by osteoblasts and hematopoietic cells. OPN suppresses the proliferation of hematopoietic stem cells in vitro and may regulate the hematopoietic stem cell pool. Increased serum OPN concentrations occur in chronic myeloid leukemia, multiple myeloma, and acute myeloid leukemia (AML). In the present study, we analyzed the prognostic impact of OPN in AML by investigating the expression and relevance of OPN in newly diagnosed AML patients from 2 large study groups (the German AML Cooperative Group and the Dutch-Belgian Hematology Oncology Cooperative group). IHC (n = 84), ELISAs of blood/BM sera (n = 41), and microarray data for mRNA levels (n = 261) were performed. Expression of OPN protein was increased in AML patients both in BM blasts (IHC) and in BM serum (ELISA) compared with healthy controls. Patients expressing high levels of OPN within the BM (IHC) experienced shortened overall survival (OS; P = .025). Multivariate analysis identified karyotype, blast clearance (day 16), and the level of OPN expression as independent prognostic factors for OS. This prompted us to analyze microarray data from 261 patients from a third cohort. The analysis confirmed OPN as a prognostic marker. In summary, high OPN mRNA expression indicated decreased event-free survival (P = .0002) and OS (P = .001). The prognostic role of OPN was most prominent in intermediate-risk AML. These data provide evidence that OPN expression is an independent prognostic factor in AML.
Introduction
Treatment strategies for patients with acute myeloid leukemia (AML) are based on various prognostic factors, including age and performance status of the patient, as well as cytogenetic and molecular characteristics of the leukemic clone. Key objectives in predicting the prognosis of AML are to define patients with high risk of relapse to apply more intensive therapy such as allogeneic stem cell transplantation, and also to withhold aggressive therapy when final success seems highly unlikely.1 Hematologic malignancies are developed and maintained not only by the molecular events inside the malignant cell clone, but also by their interaction with the microenvironment. The cross-talk between stroma and malignant cells has a major impact on pathophysiological behavior and progress of the disease.2–4
Osteopontin (OPN) is a secreted glycoprotein of the SIBLING family that is involved in physiologic and pathophysiologic processes.5 The different effects of OPN are due to its multiple receptors and isoforms. Cleavage of OPN leads to a molecular mass that facilitates binding to various receptors such as CD44, α9β1, and α4β7.5 In vitro, OPN has been shown to influence adhesion and migration of smooth muscle cells, endothelial cells, and melanoma cells.5 OPN exerts dual functions as a chemoattractant cytokine and as an extracellular component.5 Previous studies indicated a role for OPN in several malignancies. Whereas most previous studies focused on solid cancers,6,7 there have been recent advances in deciphering the role of OPN in hematologic malignancies.8–10 For example, it has been shown that OPN contributes to BM adhesion and migration of hematopoietic stem cells (HSCs).11 OPN may act as a negative regulator of HSCs through the inhibition of HSC division.11 HSC adhesion to osteoblasts leads to a resting state of these cells in the stem cell niche, with protection from cytotoxic and apoptotic stimuli.12
In the present study, we analyzed the expression of OPN in BM, AML blasts, and serum of AML patients. Our results show that the expression of OPN is increased at the mRNA and protein levels in AML patients compared with healthy controls. We also show that OPN is an independent prognostic marker in AML.
Methods
Patient characteristics
IHC analysis cohort.
BM sections from 84 patients with newly diagnosed, untreated AML were chosen randomly. Three control samples were taken from patients with various diseases but normal BM morphology. BM core biopsies were obtained at presentation. After core biopsy, BM aspiration was obtained through a separate puncture for cytological analysis. All patients were treated uniformly according to the German AML Cooperative Group (AMLCG) trial as published in detail previously.13,14 All patients gave their informed consent in accordance with the Declaration of Helsinki, and the study received approval from the local ethics committees of the participating institutions. Patient characteristics are summarized in Table 1.
Patient characteristics
| Characteristic . | IHC . | ELISA . | mRNA . |
|---|---|---|---|
| n | 84 | 41 | 261 |
| Median age, y (range) | 58 (19-82) | 61 (24-81) | < 35 y, n = 76 35-60 y, n = 177 ≥ 60 y, n =3217 |
| Sex, % | |||
| Male | 55 | 49 | * |
| Female | 45 | 51 | * |
| FAB classification (unclassified), % | 0 | 0 | 3.8 |
| M0 | 1.4 | 0 | 2.1 |
| M1 | 11.3 | 23.1 | 22.1 |
| M2 | 29.6 | 28.2 | 23.2 |
| M3 | 4.2 | 2.6 | 6.7 |
| M4 | 21.1 | 17.9 | 18.6 |
| M5 | 22.5 | 25.6 | 22.5 |
| M6 | 8.5 | 2.6 | 1 |
| M7 | 1.4 | 0 | 0 |
| Karyotype, % | |||
| Favorable: 8/21; 15/17 | 9.5 | 19.5 | 20 |
| Intermediate: normal, +8, +22, other | 37.8 | 46.3 | 47 |
| Unfavorable: complex, −5, −7, 11q | 20.3 | 9.8 | 15.1 |
| Unknown | 32.4 | 24.4 | 17.9 |
| CR following therapy, % | 64 | 69 | * |
| Median follow-up, mo (range) | 28 (1-103) | 15 (1-70) | 43 (1-213) |
| Characteristic . | IHC . | ELISA . | mRNA . |
|---|---|---|---|
| n | 84 | 41 | 261 |
| Median age, y (range) | 58 (19-82) | 61 (24-81) | < 35 y, n = 76 35-60 y, n = 177 ≥ 60 y, n =3217 |
| Sex, % | |||
| Male | 55 | 49 | * |
| Female | 45 | 51 | * |
| FAB classification (unclassified), % | 0 | 0 | 3.8 |
| M0 | 1.4 | 0 | 2.1 |
| M1 | 11.3 | 23.1 | 22.1 |
| M2 | 29.6 | 28.2 | 23.2 |
| M3 | 4.2 | 2.6 | 6.7 |
| M4 | 21.1 | 17.9 | 18.6 |
| M5 | 22.5 | 25.6 | 22.5 |
| M6 | 8.5 | 2.6 | 1 |
| M7 | 1.4 | 0 | 0 |
| Karyotype, % | |||
| Favorable: 8/21; 15/17 | 9.5 | 19.5 | 20 |
| Intermediate: normal, +8, +22, other | 37.8 | 46.3 | 47 |
| Unfavorable: complex, −5, −7, 11q | 20.3 | 9.8 | 15.1 |
| Unknown | 32.4 | 24.4 | 17.9 |
| CR following therapy, % | 64 | 69 | * |
| Median follow-up, mo (range) | 28 (1-103) | 15 (1-70) | 43 (1-213) |
Not known.
ELISA cohort.
BM serum of 41 patients with newly diagnosed, untreated AML were chosen randomly. BM serum from 4 healthy controls was also obtained. Serum was obtained by standard procedures. In addition, serum levels of OPN of the same patients were measured in peripheral blood samples. Blood samples were obtained at the time of diagnosis before the initiation of chemotherapy and stored at −80°C. All patients were treated according to the AMLCG protocol.13,14
Microarray-based analysis of mRNA levels.
Patients were treated according to the protocols of the Dutch-Belgian Hematology Oncology Cooperative group (HOVON; available at www.hovon.nl).15–17 A total of 284 AML patients provided BM aspirates or peripheral blood samples at the time of diagnosis. Patients had received a diagnosis of primary AML, which had been confirmed by a cytologic examination of blood and BM (Table 1). Blasts and mononuclear cells were purified by Ficoll-Hypaque (Nygaard) centrifugation and cryopreserved as described previously.18 Patient samples were analyzed with U133A GeneChips (Affymetrix). The OPN level could be measured in 261 patients. Further details have been described previously.18
IHC
IHC staining.
BM specimens were fixed in paraformaldehyde, decalcified, and embedded in paraffin. The anti–human OPN Ab (AF1433; R&D Systems) was used for staining, which was performed using the alkaline phosphatase/antialkaline phosphatase double-bridge technique (APAAP kit; Dako). Briefly, tissue sections were deparaffinized and rehydrated. Samples were pretreated to promote antigen retrieval in a microwave oven in 10mM sodium citrate. The primary Abs were applied overnight. Subsequent steps were performed according to the manufacturer's instructions. Sections were counterstained with Erythrosin solution.
IHC expression.
OPN expression was semiquantitatively assessed by scoring the proportion and intensity of stained cells according to our previously published methods.19 The entire BM section was scanned at low magnification and the percentage of positive cells stained was estimated according to a 3-grade scale. The intensity was subsequently evaluated in 3 representative 500× fields. The mean of cellular staining intensity was subsequently multiplied by the number of positive cells according to the 3-grade scale and results are expressed as arbitrary units (AUs). Expression was evaluated in 2-3 sections processed in independent immunostainings and the mean value was calculated.
ELISAs
ELISAs were performed using kits from R&D Systems. Briefly, patient samples were collected using EDTA and stored at −80°C. Serum samples were added to separate microplates containing specific Abs. Mixtures were incubated at room temperature. Plates were washed to remove unbound antigen. Enzyme-linked Abs specific for OPN were then added and the mixtures were incubated and washed. The substrate solution was added to the wells. Color development was stopped and the intensity was measured. Optical density of each well was determined according to the manufacturer's instructions.
Statistics
Beyond descriptive statistical analyses, inferential analyses were performed using nonparametric methods. Differences in OPN expression between 2 groups were analyzed using the Mann-Whitney U test for independent groups. In the case of more than 2 groups, the Kruskal-Wallis test was used. Correlation between metric variables was assessed by the Spearman rank correlation coefficient (rs). End points were defined as described previously.20 Patients who survived the entire follow-up period were censored at the date of last contact. Cutoff values of OPN levels were determined by recursive partitioning. In addition, bootstrap analysis and hazard ratio (HR) per unit were determined to identify the stability of the identified cutoff points. Survival curves were generated using the Kaplan-Meier method and the log-rank test was used to compare survival between groups. Univariate and multivariate Cox proportional hazards-regression analyses were performed to evaluate the impact of prognostic factors on survival. Factors found to be statistically significant at a 10% level in the univariate analyses were included in the multivariate model. Two-sided P ≤ .05 was considered to indicate statistical significance. All calculations were performed using SPSS Version 18 software.
Results
IHC analysis
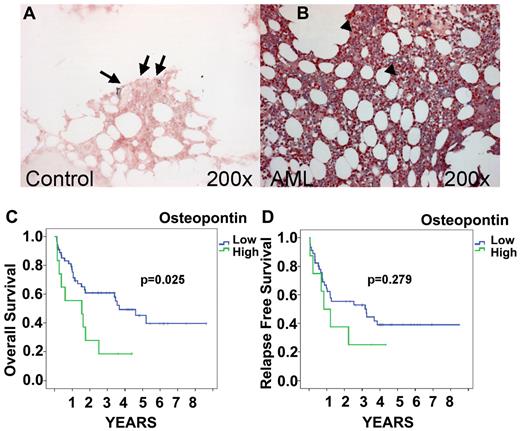
Expression of OPN was analyzed by IHC staining in BM core biopsies of 84 AML patients. Patient characteristics are provided in Table 1. In AML and control patients, staining intensity varied from weak to moderate, with some scattered cells showing strongly positive signals (Figure 1). OPN-positive leukemic blasts were widely and uniformly observed in a homogeneous pattern throughout the cellular regions of AML BM. Signals were associated with the cytoplasm and the nucleus of leukemic blasts. In contrast to its expression in AML, OPN expression in control BM samples was generally weak and limited to a few scattered positive cells in the endosteal region, with most of the cells being completely negative (Figure 1A). The staining scores for OPN protein expression in AML patients showed a median of 6.27 AU (range, 1-14) and differed significantly from the OPN expression found in BM sections of controls (1.33 AUs; range, 1.3-2.0; P = .0059 by Mann-Whitney test). To verify the concordance of the 3 observers, we performed a reliability analysis. The intraclass-correlation analysis revealed good reliability (0.654; 95% confidence interval [CI], 0.536-0.753). No significant association was found between the expression of OPN and age at diagnosis or sex. Although OPN was detected consistently in leukemic blasts, staining scores were not associated with the percentage of leukemic blast infiltration. We then analyzed the correlation between OPN expression and AML subtypes according to French-American-British (FAB) classification as defined by cytological analysis. Statistical analyses did not reveal differences in expression between subtypes M0 to M7 (P = .614 by Kruskal-Wallis test). In addition, no association was detected between the level of expression of OPN and the response to therapy at day 16 (P > .5 for each variable).
OPN protein expression by IHC and survival. OPN expression in control BM (A) and AML (FAB AML M5; B) obtained at presentation. IHC localization was performed using the respective specific Abs and the alkaline phosphatase/antialkaline phosphatase technique. Note the expression of OPN in the BM sections of AML patients (B arrowhead) compared with the controls (A arrow). Original magnification, 200×. Images were captured with Axiocam (Zeiss) using Axiovision (Zeiss) and Image-Pro Plus (Media Cybernetics). Kaplan-Meier curves of OS and relapse-free survival for AML patients stratified for BM OPN expression are shown. OS of patients expressing high levels of OPN (≥ 9.662 AUs) in the BM was significantly lower than of patients with low OPN expression (< 9.662 AUs; P = .025; C). Although a visible trend, the difference found for relapse-free survival was not significant (P = .279; D).
OPN protein expression by IHC and survival. OPN expression in control BM (A) and AML (FAB AML M5; B) obtained at presentation. IHC localization was performed using the respective specific Abs and the alkaline phosphatase/antialkaline phosphatase technique. Note the expression of OPN in the BM sections of AML patients (B arrowhead) compared with the controls (A arrow). Original magnification, 200×. Images were captured with Axiocam (Zeiss) using Axiovision (Zeiss) and Image-Pro Plus (Media Cybernetics). Kaplan-Meier curves of OS and relapse-free survival for AML patients stratified for BM OPN expression are shown. OS of patients expressing high levels of OPN (≥ 9.662 AUs) in the BM was significantly lower than of patients with low OPN expression (< 9.662 AUs; P = .025; C). Although a visible trend, the difference found for relapse-free survival was not significant (P = .279; D).
We also analyzed the association between pretherapeutic OPN expression and prognosis in AML (Figure 1C-D). Optimized cutoff points were identified by recursive partitioning, and an AU of 9.662 was defined as the cutoff.20 A total of 18.2% of the analyzed AML patients had an OPN expression level above that cutoff value. Univariate analysis revealed that patients expressing high levels of OPN (AU > 9.662) in the BM had significantly shorter OS than those with low OPN levels (P = .025). The median survival time of high OPN–expressing AML patients was more than 2-fold longer than survival time of patients with low OPN expression (3.7 vs 1.5 years, respectively). Figure 1 shows the OS stratified according to OPN expression. A multivariate Cox-regression analysis was performed that included the parameters FAB subtype, karyotype, and OPN expression. OPN emerged as an independent prognostic factor for OS (P = .03; HR = 2.35; 95% CI, 1.09-5.09). In addition, we observed a trend between high OPN expression and relapse-free survival (Figure 1D), although this correlation failed to reach the level of significance (P = .279), most likely because of the lower number of patients available for this analysis.
OPN levels in serum
Levels of OPN were analyzed in pretherapeutic BM and blood serum samples from patients with AML and from healthy subjects (Table 1). The median level of OPN in AML patients was 5.4 ng/mL (range, 0.3-20.0). In blood serum of AML patients, the levels of OPN were increased significantly compared with controls. Blood OPN protein levels in AML patients were at a median of 7.4 ng/mL (range, 0.3-30.5) and differed significantly from OPN levels found in blood serum of controls (median, 2.1 ng/mL; range, 0.83-3.22; P = .01 by Mann-Whitney test). No statistical association between the levels of OPN in blood and BM and the variables sex and age at diagnosis of AML were observed. Levels of OPN did not differ significantly between the different AML subtypes as defined by the FAB classification (P > .1 for each variable). Furthermore, OPN levels did not differ significantly among cytogenetic risk groups.
The BM of the AML patients studied was highly infiltrated by leukemic blasts. The median percentage of blasts was 70% (range, 15%-99%). The ELISA levels of OPN were not correlated positively with the percentage of leukemic blast infiltration as determined by BM aspiration. In addition, OPN was not associated with WBC count or lactate dehydrogenase levels. We also investigated whether levels of OPN at first diagnosis could predict remission after induction therapy. Of the 41 patients in our study population receiving intensive induction therapy, 78% achieved a complete remission (CR). Levels of OPN in these patients did not differ significantly from those observed in patients not achieving a CR (P = .280).
To determine the relationship between OPN protein expression levels and prognosis in AML, we performed univariate Cox-proportional hazard analyses. As expected, the variables karyotype (favorable vs intermediate/unfavorable), WBC, and lactate dehydrogenase displayed prognostic significance. Furthermore, using the dichotomous cutoff levels identified by recursive partitioning, OPN protein expression in the BM was significantly associated with survival (Figure 2A-B). Blood OPN levels showed no prognostic significance (data not shown). OS and event-free survival (EFS) were significantly lower for patients with BM OPN levels above 6.32 ng/mL in the BM serum.
BM serum OPN expression by ELISA. Kaplan-Meier curves for OS (A) and EFS (B) of AML patients stratified for BM serum OPN expression. OS of patients with low (< 6.632 ng/mL) levels of OPN in the BM was significantly higher than in patients with high (≥ 6.632 ng/mL) OPN expression (P = .018; A). Similar differences were found for EFS (P = .008; B).
BM serum OPN expression by ELISA. Kaplan-Meier curves for OS (A) and EFS (B) of AML patients stratified for BM serum OPN expression. OS of patients with low (< 6.632 ng/mL) levels of OPN in the BM was significantly higher than in patients with high (≥ 6.632 ng/mL) OPN expression (P = .018; A). Similar differences were found for EFS (P = .008; B).
OPN is increased in AML blasts at the mRNA level
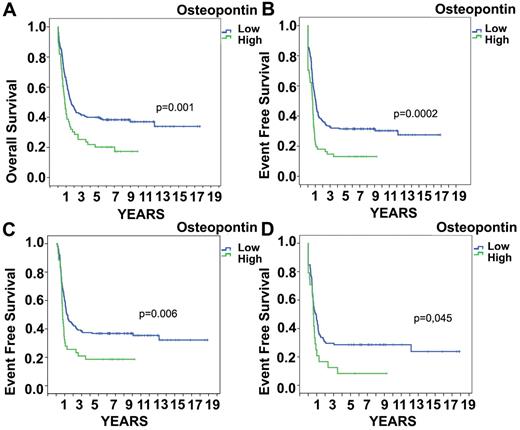
To confirm the prognostic significance of OPN in AML, we analyzed a published microarray analysis in AML patients from the HOVON group using the database published by Valk et al.18 OPN was expressed at a low signal-to-noise ratio and the specific queries concerning OPN levels could be analyzed. The median OPN mRNA expression level in these AML patients was 4.9 (range, 4.18-9.27). OPN levels were increased significantly in the subgroup of AML M3, whereas it did not differ significantly between the other different AML subtypes as defined by the FAB classification (P > .1 for each variable by Kruskal-Wallis test). In the subgroup of AML M3, OPN expression had no significant impact on EFS/OS. In addition, OPN levels did not differ significantly among cytogenetic risk groups. To determine the relationship between OPN and prognosis in AML, we initially performed univariate Cox proportional-hazards analyses. An optimal cutoff value of OPN level was determined by recursive partitioning. As expected, the variables karyotype and molecular genetics displayed prognostic significance.18 OPN was also significantly associated with survival (Figure 3). The HR of death was significantly higher for patients with high OPN levels (HR = 1.721; 95% CI, 1.24-2.40; P = .001).
OPN mRNA expression and survival. High OPN expression on the mRNA level is associated with significantly (P = .001) shorter OS (A) and EFS (P = .0002; B). In the group of AML patients who reached remission, increased OPN expression still showed significantly shorter EFS (P = .006; C) and this was also true in intermediate-risk patients (as defined by cytogenetics; P = .045; D).
OPN mRNA expression and survival. High OPN expression on the mRNA level is associated with significantly (P = .001) shorter OS (A) and EFS (P = .0002; B). In the group of AML patients who reached remission, increased OPN expression still showed significantly shorter EFS (P = .006; C) and this was also true in intermediate-risk patients (as defined by cytogenetics; P = .045; D).
We also performed multivariate Cox-regression analyses incorporating all variables showing a significant association with survival in the univariate model at the 10% level. The multivariate analysis of OPN mRNA included the covariates FLT3, CEBPA, FAB type, and cytogenetics. OPN emerged as an independent predictor of survival (HR = 1.708; 95% CI, 1.141-2.557; P = .009; supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Figure 3 shows the Kaplan-Meier curves of OS and EFS stratified for OPN levels. An association was detected between high levels of OPN expression and resistance to induction therapy (P = .009). We could only speculate that OPN may act at that early time point as a negative regulator of leukemic cell proliferation and thus chemotherapy resistance. This prompted us to investigate whether OPN expression levels at diagnosis would influence outcome even after an initial remission was reached. The analysis was restricted to patients who achieved a CR after induction chemotherapy. Nonetheless, OPN expression levels at diagnosis showed a significant and independent impact on survival (Figure 3C; P = .016). Further, we analyzed OPN levels in subsequently relapsing patients compared with patients with continuous CR and found significantly more high OPN–expressing blasts (12% vs 27%; P < .012) in patients who subsequently relapsed. In the patient group who died in CR (n = 36), the numbers were too low for statistical analysis. Only 9 patients were in the high OPN group. In addition, we analyzed whether OS and EFS were altered by exclusion of AML M3 patients, who have a higher expression of OPN in contrast to the other FAB types. The significance value for OS was not altered, whereas, as expected, the EFS was even more significant; the P value changed from .0002 to .00003. For patients who reached CR, high OPN expression was still significant in terms of EFS (P = .006). We subsequently investigated whether OPN presents a prognostic factor in the different cytogenetic risk groups. Interestingly, OPN remained a significant prognostic factor in intermediate-risk patients (Figure 3D; P = .045).
Validation of cutoff values and HR per unit increase in OPN
To validate the determined optimal cutoff values of OPN levels, a bootstrap approach was applied. The 1000 bootstrap sample results showed weak reproducibility (supplemental Table 2). This finding indicates that the cutoff points used in our analyses should not be regarded as the best that can be achieved in different experiments. Rather, our findings indicate that rising OPN levels indicate increased risk for therapy failure. In all 3 analyses, OPN levels showed a unfavorable HR per unit increase in OPN that reached significance in the case of mRNA OPN (supplemental Table 3).
Discussion
In this retrospective study, we identified OPN expression as analyzed on the mRNA and protein level as a prognostic marker in AML. In 3 different patient cohorts and with various methods, we demonstrated that increased expression of OPN was associated with shortened survival. Whereas in control BM, OPN was only expressed by osteoblasts within the endosteal region, we could demonstrate that OPN is readily expressed in AML blasts. The analyses (cutoff value defined by statistical optimization) revealed that OPN is highly expressed in approximately 20% of AML patients (ELISA, 23%; IHC, 18%; RNA, 23%) and is neither associated with FAB type, nor with cytogenetic or molecular alterations. In addition, in a recently published dataset from the MILE study, we also identified significantly increased levels of OPN in BM from AML patients compared with normal BM (data not shown).21 Several studies have indicated a role for OPN in different malignancies. Whereas most of the publications had focused on solid cancers,6,7 there have been recent advances in deciphering the role of OPN in hematologic malignancies.8–10 Increased serum OPN concentrations have been reported in chronic myeloid leukemias, multiple myelomas, and AML.22 Lee et al demonstrated in a small cohort that BM levels of higher OPN were associated with significant shorter survival times in AML patients in a 1-year survival analysis, whereas 5-year analysis revealed no statistical difference.7 Powell et al23 performed expression screening to identify targets in AML. These analyses identified OPN as a functionally relevant target. siRNA-mediated knockdown of OPN expression induced cell death in both AML blasts and leukemic progenitors. That study demonstrated that in normal karyotype AML, increased expression of OPN at diagnosis is associated with poor prognosis (n = 60).23 The important function of OPN in normal hematopoiesis appears to be the suppression of proliferation of primitive stem cells in the BM niche, as evidenced in OPN−/− mice and in vitro colony formation assays.11,24 These data therefore provide strong evidence that OPN is an important component of the HSC niche, participating in HSC location and as a physiologically negative regulator of HSC proliferation.11 Furthermore, exogenous OPN suppresses the proliferation of primitive HSCs in vitro. The relevance of this observation is demonstrated by the markedly enhanced cycling of HSC in OPN−/− mice11 and their hypersensitivity to exogenous stimuli.24,25 Considering these biologic effects of OPN and the significant influence of OPN expression on prognosis as measured by EFS and OS in our present study, we assume that OPN contributes to chemotherapy resistance in a paracrine/autocrine manner.
The self-renewal of leukemic stem cells (LSCs) may have an extrinsic component like OPN as in normal HSCs.24 The importance of the LSC microenvironment has also been demonstrated previously.26 These data showed that mesenchymal stem cells are essential for the long-term survival and expansion of leukemic lymphoblasts in childhood acute lymphoblastic leukemia.26 The microenvironment supports the repopulating potential and the ability to propagate and maintain the leukemic phenotype. Many studies have reported that AML LSCs are quiescent or slowly dividing, whereas clonogenic progenitors are rapidly proliferating.27–29 However, there is emerging evidence that the complex interactions between LSCs and their niche may also be targeted. Interestingly, targeting of CD44, a known ligand of OPN, eradicates human myeloid leukemic stem cells in mice.30 In summary, the results of the present study suggest that OPN, a molecule important for stem-cell regulation, is closely associated with patient prognosis in AML. Further studies should investigate OPN as a biomarker and a possible target for AML therapy.
The online version of this article contains a data supplement.
Data reported herein were presented in part at the annual meeting of the German Society for Hematology and Oncology; October 4, 2010; Berlin, Germany.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Peter J. Valk and Ruud Delwel (Department of Hematology, Erasmus University Medical Center, Rotterdam, The Netherlands) for their review and helpful discussions and H. Hintelmann and R. Roß (Department of Medicine A, University Hospital, Muenster, Germany) for expert technical assistance.
Authorship
Contribution: R.L., T.K., W.E.B., and R.M. conceived and designed the study; R.L., C.S., C.B., I.A., T.K., B.L., T.B., W.H., C.M.-T., W.E.B., and R.M. provided study materials or patients; R.L., J.G., C.S., M.B., C.S., T.K., B.L., T.B., W.H., C.M.-T., W.E.B., and R.M. collected and assembled the data; R.L., J.G., C.S., M.B., C.S., C.B., B.L., T.B., W.H., C.M.-T., W.E.B., and R.M. analyzed and interpreted the data; R.L., C.M.-T., W.E.B., and R.M. wrote the manuscript; and all authors approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ruediger Liersch, MD, Department of Medicine A–Hematology and Oncology, University of Muenster, Albert-Schweitzer-Campus 1a, D-48129 Muenster, Germany; e-mail: rliersch@uni-muenster.de.
References
Author notes
C.M.-T., W.E.B., and R.M. contributed equally to this work and share senior authorship.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal