Plasmacytoid dendritic cells (pDCs) selectively express Toll-like receptor (TLR)–7 and TLR-9, which allow them to rapidly secrete massive amounts of type I interferons after sensing nucleic acids derived from viruses or bacteria. It is not completely understood how development and function of pDCs are controlled at the transcriptional level. One of the main factors driving pDC development is the ETS factor Spi-B, but little is known about its target genes. Here we demonstrate that Spi-B is crucial for the differentiation of hematopoietic progenitor cells into pDCs by controlling survival of pDCs and its progenitors. In search for Spi-B target genes, we identified the antiapoptotic gene Bcl2-A1 as a specific and direct target gene, thereby consolidating the critical role of Spi-B in cell survival.
Introduction
Plasmacytoid dendritic cells (pDCs) form a unique subset within the DC lineage. In contrast to conventional dendritic cells (cDCs), pDCs express Toll-like receptor (TLR)–7 and TLR-9, which recognize viral and microbial single-stranded RNA or double-stranded DNA, respectively (reviewed in Liu1 ). TLR activation in pDCs leads to rapid secretion of high amounts of type I interferons (IFNs), which initiate antiviral immune responses. In addition, pDCs mature in response to autocrine production of proinflammatory cytokines such as IL-6 and tumor necrosis factor (TNF)–α. Collectively, this contributes to activation of T, B, and NK cells (reviewed in Lande and Gilliet2 ). pDCs originate from hematopoietic progenitor cells (HPCs) in the bone marrow, but can also develop in the thymus.3 Although both myeloid and lymphoid precursors give rise to pDCs, myeloid derivation is predominant (reviewed by Naik4 ) and depends on Fms-like kinase 3 ligand (Flt3L).5
The ETS family member Spi-B is required for pDC development, because both in vitro and in vivo human HPCs failed to give rise to pDCs when inhibiting Spi-B expression.6 Spi-B shares with other ETS members a conserved ETS domain that mediates DNA binding.7 Spi-B is a hematopoietic cell–specific transcription factor8 and is expressed, in addition to pDCs, in CD34+ HPCs,9 pro-T cells,10 and mature B cells.9 Spi-B potently regulates lineage commitment during human hematopoiesis as its overexpression in HPCs blocks T, B, and NK cell development, but promotes pDC development.6 Despite the importance of Spi-B in development of immune cells, little is known about its direct target genes. In mice, Spi-B may act by indirectly promoting the activity of the E-protein E2-2, which both in human and mouse is critical for pDC development.11,12 In human B cells, ectopic expression of Spi-B blocked memory B-cell differentiation into antibody secreting cells by direct transcriptional repression of the PRDM1 and XBP1 genes.13
pDCs are fragile cells that when cultured in vitro rapidly undergo apoptosis, which can be counteracted by stimulation with TLR-7/9 agonists or cytokines, including IL-3, granulocyte macrophage–colony stimulating factor (GM-CSF), or IFN-α.14 Apoptosis is a highly regulated process that serves to remove superfluous, damaged, or infected cells and therefore to maintain cellular homeostasis (reviewed by Elmore15 ). Apoptosis and cell survival are intimately linked, because their regulation is based on the balance between proapoptotic and antiapoptotic (survival) regulators. Members of the Bcl-2 family regulate the intrinsic pathway of apoptosis through control of the outer mitochondrial membrane (OMM) integrity. Proapoptotic members such as Bak, Bax, Bim, and Noxa share a so-called Bcl-2 homology domain, which promote OMM permeability leading to the release of cytochrome C in the cytoplasm and apoptosis induction. Antiapoptotic members, including Bcl-2, Bcl-xL, Bcl-w, Bcl2-A1, and Mcl-1, promote cell survival through counteracting proapoptotic Bcl-2 member activity.
Bcl2-A1 is a hematopoietic specific protein and protects cells from apoptosis induced by a variety of apoptotic stimuli, such as death-receptor ligation,16 p53 overexpression,17 and DNA-damaging agents.18 In mice, Bcl2-A1 is expressed in a variety of hematopoietic cell lineages, including T-helper lymphocytes, macrophages, and neutrophils.19 In humans, Bcl2-A1 is expressed in various types of hematopoietic cells in the bone marrow and fetal liver (FL), and in germinal centres of peripheral lymphoid organs.20 The BCL2-A1 gene is induced in response to GM-CSF21 and is a direct transcriptional target of nuclear factor (NF)–κB in response to inflammatory mediators.22,–24
To investigate whether Spi-B has a role in regulating pDC survival and/or proliferation, we set out to use the leukemic pDC cell line CAL-125 as a model to study gene regulation in pDCs. We studied the gene expression profile of this cell line under conditions of overexpression and inhibition of Spi-B. Using this approach, we identified BCL2-A1 as a direct target of Spi-B. We validated and confirmed our findings in primary pDCs, which underlined the role of Spi-B in pDC development and survival through regulation of the antiapoptotic gene BCL2-A1.
Methods
Cell lines
The pDC cell line CAL-125 was cultured in RPMI-1640 medium (Invitrogen) supplemented with 8% FCS, and maintained at 37°C, 5% CO2.
Reagents
Cells were activated with 10 ng/mL IL-3 (R&D Systems) in the presence of CD40 ligand–transfected L cells (10 000/well, irradiated at 7000 rads) in RPMI-1640 medium (Invitrogen), 8% FCS. CpG-A (ODN2216), CpG-B (ODN2006), and R848 were purchased from Invivogen. HSV-1 (KOS strain; attenuated by γ irradiation; gift from R. Chase, Schering-Plough, Kenilworth, NJ) was added at 10 PFU/cell.
Isolation of CD34+ cells from FL
Human FLs were obtained from elective abortions. Gestational age was determined by ultrasonic measurement of the skull diameter and ranged from 14 to 20 weeks. The use of fetal tissue was approved by the Medical Ethical Committee of the Academic Medical Center and was contingent on obtaining informed consent, in accordance with the Declaration of Helsinki. For purification of CD34+ cells, FL cells were isolated from a Ficoll-Hypaque density gradient (Lymphoprep; Nycomed Pharma). Subsequently, CD34+ cells were enriched by immunomagnetic cell sorting, using a CD34+ separation kit (Miltenyi Biotec). CD34+CD38− FL hematopoietic progenitors were sorted to purity using a fluorescence-activated cell sorter (FACS) Aria flowcytometer (BD Bioscience) after labeling with fluorescent conjugated antibodies. Purity was ≥ 99% and confirmed by reanalysis of sorted cells.
Isolation of primary human pDC from thymus
Postnatal thymic (PNT) tissue was obtained from surgical specimens removed from children up to 3 years of age undergoing open-heart surgery (Leids Universitair Medisch Centrum, Leiden, The Netherlands), approved by the Medical Ethical Committee of the Academic Medical Center and in accordance with the Declaration of Helsinki. Thymocytes were isolated from a Ficoll-Hypaque density gradient. Subsequently, BDCA4+ cells were enriched by immunomagnetic bead selection using a BDCA4-cell separation kit (Miltenyi Biotec). CD123+CD45RA+ pDCs were sorted by flow cytometry on a FACSAria (BD Bioscience) after labeling with fluorescent conjugated antibodies.
Flow cytometry
FL progenitor cells were sorted using CD34-PE, CD38–PE-Cy7, and lineage markers-APC (CD3, CD14, CD19, CD56, BD Bioscience; BDCA2, Miltenyi Biotech). Thymic progenitor cells were sorted using CD34–PE-Cy7 and CD1a-PE (BD Bioscience). For analysis, single cell suspensions were stained with fluorescein isothiocyanate (FITC), phycoerythrin (PE), PE-Cy7, allophycocyanin (APC), or APC-Cy7 coupled antihuman monoclonal antibodies targeting the following cell surface markers: CCR7, CD40, CD45RA, CD56, CD62L, CD80, CD86, CD123, HLA-DR (BD Bioscience), and BDCA2 (Miltenyi Biotech). Samples were analyzed on a LSRII (BD Bioscience) and analyzed using FlowJo Version 7 software (TreeStar). Cell proliferation was assessed using the CellTrace-violet proliferation kit (Invitrogen) according to the manufacturer's instructions. For apoptosis staining we used annexin V-PE (BD Bioscience) and 7-AAD viability staining solution (Ebioscience).
Retroviral and lentiviral constructs and transductions
For overexpression of Spi-B we used either the retroviral construct pLZRS–Spi-B–IRES-green fluorescent protein (GFP) or pLZRS–Spi-B∼ER-IRES-GFP.26 ΔEts and Δ transactivation domain (ΔTAD) truncated version of Spi-B were generated as previously described.13 To knockdown Spi-B in CAL-1 cells, Spi-B short hairpin (sh)RNA was cloned from a retroviral backbone6 into a lentiviral backbone (pTRIP-H1–Spi-B shRNA/EF1α-GFP). For virus production, constructs were transfected into the Phoenix-GalV packaging cells (retroviral) or 293T cells (lentiviral).26,27 Control cells were transduced with empty pLZRS-IRES-GFP constructs or pTRIP expressing an irrelevant shRNA targeting renilla mRNA (pTRIP-H1-renilla shRNA/EF1α-GFP).6 For transduction of CAL-1 cells, 106 cells were transferred to plates coated with retronectin (30 μg/mL, Takara) and incubated with virus supernatants for 6 hours. To induce nuclear translocation of estrogen receptor (ER) tagged Spi-B, cells were treated with 0.5μM 4-hydroxytamoxifen (4HT; Sigma-Aldrich). De novo protein translation was inhibited by preincubation with 2.8 μg/mL cycloheximide (CHX; Sigma-Aldrich). For Bcl2-A1 knockdown experiments, shRNAs specifically targeting the Bcl2-A1 mRNA were designed using Ambion's siRNA Target Finder (http://www.ambion.com) and subcloned into pRETRO-pgk-GFP.6
PCR
Total RNA was extracted using Trizol reagent (Invitrogen). RNA concentration and quality were determined using the Nanodrop spectrophotometer. Equal amounts of total RNA were reverse transcribed into cDNA with first strand buffer, superscript II reverse transcriptase (Invitrogen), dNTPs (Roche), and Oligo (dT; Promega), or using the RNA-to-cDNA kit (Roche). cDNA was amplified using a polymerase chain reaction (PCR) machine for conventional RT-PCR and separated on a 1.5% agarose gel, or amplified using an iCycler and SYBR green supermix (BioRad) for quantitative PCR (qPCR) using specific primer sets (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Each sample was analyzed in triplicates and expression levels were normalized to the 3 housekeeping genes HPRT, GAPDH, and 18S RNA.
Chromatin immunoprecipitation assay
Spi-B∼ER-GFP+ cells (1 × 107) were incubated with or without 4HT for 4 hours. Chromatin immunoprecipitation (ChIP) was performed according to an adapted version of the Upstate ChIP kit protocol (Upstate BiotechnologyA) with either 3 μg polyclonal anti-ER antibody (Santa Cruz Biotechnology) or 3 μg normal rabbit immunoglobulin (Ig)G (Invitrogen). Precipitated chromatin was purified with a PCR purification kit (QIAGEN) and analyzed by QPCR using 5 different primer sets, each of them specific for a different DNA sequence located within the BCL2-A1 promoter region up to 3.2 Kb upstream of the transcription start (PR1-PR5; supplemental Table 1). As a positive control, the CD40 promoter was amplified by qPCR using the primer set PR+.
Luciferase reporter assays
Reporter constructs were generated using PCR to amplify a 3.2 Kb DNA fragment containing the conserved human Bcl2-A1 promoter region (full-length) and a truncated version of the Bcl2-A1 promoter region (Δ5′; Figure 4C). Amplified PCR fragments were cloned into the pCR2-1TOPO TA vector (Invitrogen). Sequencing with the primer sets used for ChIP assays was performed using an ABI sequencer (Perkin Elmer) with the dye-terminator cycle-sequencing kit (Perkin Elmer). The inserts were subcloned into pGL3 basic vector (Promega) using XhoI-HindIII restriction sites. Cotransfection of one of the reporter constructs, the constitutive active renilla reniformis luciferase-producing vector prL-CMV (Promega), and pLZRS expressing different variants of Spi-B, or PU.1 as a positive control, in 293T cells was done with the Fugene transfection reagent (Roche). Detection of firefly and Renilla reniformis luciferase was done using the Dual Luciferase assay kit (Promega) on a Synergy HT microplate reader (Biotek).
Cytometric bead array analysis and ELISA
CAL-1 cells were stimulated for 6 hours with CpG-B (ODN2006; 12.5 μg/mL) or R848 (10 μg/mL). Cell-free supernatants were collected and analyzed for cytokine content using the Cytometric Bead Array (CBA), according to the manufacturer's protocol (CBA Human Inflammation Kit; BD Bioscience). Quantification of IFN-β levels in supernatants was done using an enzyme-linked immunosorbent assay (ELISA) kit (PBL Interferon Source) according to the manufacturer's instructions.
In vivo growth assay
Briefly, 0.25 × 106 CAL-1 cells transduced with Spi-B shRNA/EF1α-GFP or renilla shRNA/EF1α-GFP control were subcutaneously injected into the right and left flank of RAG-2−/−γc−/− mice.28 To mimic a tumor like growth, cells were mixed with collagen (Matrigel; Invitrogen) before injection. Twenty days after engraftment mice were killed and tumors isolated. Cells were counted and the percentage of GFP+ cells was analyzed by flow cytometry. Different organs of the mice were analyzed (blood, spleen, liver) for the presence of GFP+ cells.
In vitro differentiation assay
Sorted CD34+CD38− HPCs from FLs were transduced as previously described.26 Briefly, transduced progenitors were cultured on a layer of mouse OP9 stromal cells in the presence of 5 ng/mL Flt3L and 5 ng/mL IL-7 (PeproTech). In vitro generated GFP+CD123hiBDCA2+ pDCs were analyzed by flow cytometry after 7 days of coculture. The fold expansion in absolute cell numbers was calculated on basis of total numbers of cells harvested from the cultures, percentages of transduced cells, and percentages of each population corrected for the number of input cells.
Statistical analyses
Data were subjected to 2-tailed paired Student t test analysis using Prism 5 for Windows (Graphpad) and considered significant when P values were at least < .05.
Results
CAL-1 cells represent a valid model to study certain aspects of primary pDCs
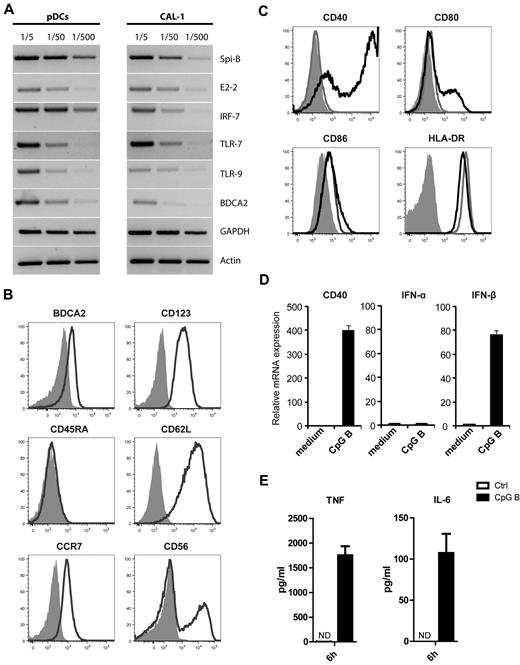
As it is technically challenging to study molecular mechanisms involved in differentiation and activation of fresh human pDCs, we made use of the previously established pDC cell line CAL-1 derived from a patient with CD4+CD56+ hematodermic neoplasm.25 To further characterize the CAL-1 cell line we first measured expression of genes known to be involved in development and activation of human pDCs by semiquantitative RT-PCR (Figure 1A) and compared expression with that in primary pDCs. Both freshly isolated pDCs and CAL-1 cells expressed the ETS transcription factor Spi-B, the basis-helix-loop-helix factor E2-2, the interferon response factor IRF-7, TLR-7 and TLR-9, and the pDC restricted C-type lectin BDCA2. Except for CD45RA, the surface markers CD123 and BDCA2, as well as CCR7 and CD62L, were detected by FACS analysis (Figure 1B). In accordance with earlier observations, a fraction of CAL-1 cells expressed CD56.25 To validate this cell line as an accurate model to investigate pathways involved in pDC maturation and function, we evaluated the capacity of CAL-1 cells to respond to various stimuli, including the TLR-9 agonist CpG-B ODN, the TLR-7 ligand R848, CD40L plus IL-3, and virus (HSV-1; Figure 1C-E, data not shown). Overnight stimulation of CAL-1 cells with CpG-B strongly increased the surface expression of maturation markers, including CD40, CD80, and to a lesser extent CD86 (Figure 1C). HLA-DR was already highly expressed and not further up-regulated after activation. In accordance with these results, CD40 mRNA expression was strongly up-regulated within 4 hours of stimulation (Figure 1D). In addition, CpG-B–induced TLR-9 triggering induced IFN-β mRNA (Figure 1D) and protein (supplemental Figure 1) expression. CAL-1 cells stimulated with CpG-B either for 4 hours or overnight did not express IFN-α as determined by PCR, neither with primers recognizing specific IFN-α mRNA subtypes nor with primers recognizing all IFN-α subtypes (Figure 1D, data not shown). As previously reported,25 also CpG-A did not induce IFN-α under these conditions. CAL-1 cells stimulated with CpG-B for 6 hours produced the proinflammatory cytokines IL-6 and TNF-α as detected by cytokine-bead-array experiments (Figure 1E). Stimulation with the TLR-7 agonist R848 similarly induced expression of these cytokines, although to lesser degree than CpG-B (supplemental Figure 1). Notably, CAL-1 cells did not produce any proinflammatory cytokines in the absence of stimulation. Taken together, these data provide convincing evidence that CAL-1 cells share common genetic and functional features with primary pDCs. Therefore, we consider CAL-1 cells a valid model to study molecular mechanisms involved in pDC activation and maturation.
CAL-1 cells closely resemble primary pDCs. (A) The leukemic pDC cell line CAL-1 expresses transcripts commonly present in freshly isolated primary pDCs, including Spi-B, E2-2, IRF-7, TLR-7, TLR-9, and BDCA2 as shown by semiquantitative RT-PCR. The housekeeping genes GAPDH and actin are shown as loading controls. (B) Flow cytometric analysis of CAL-1 cells after staining with antibodies directed against the cell surface markers BDCA2, CD123, CCR7, CD45RA, CD62L, and CD56 (black lines). Isotype control stainings are shown as gray filled histograms. (C) Surface expression of the costimulatory molecules CD40, CD80, CD86, and HLA-DR was measured by flow cytometry on unstimulated CAL-1 (gray lines) and after 20 hours stimulation with CpG-B (black lines; isotype control stainings are shown as gray filled histograms). (D) CAL-1 cells were stimulated for 4 hours with CpG-B or left unstimulated in medium only, and gene expression levels of CD40, IFN-α and IFN-β1 were measured in stimulated versus unstimulated cells by qPCR. (E) CAL-1 cells were cultured in the presence of CpG-B or medium for 6 hours. Culture supernatants were analyzed for the presence of TNF-α and IL-6 by cytokine bead array analysis. ND indicates not detectable (below detection sensitivity of the assay of 4 pg/mL).
CAL-1 cells closely resemble primary pDCs. (A) The leukemic pDC cell line CAL-1 expresses transcripts commonly present in freshly isolated primary pDCs, including Spi-B, E2-2, IRF-7, TLR-7, TLR-9, and BDCA2 as shown by semiquantitative RT-PCR. The housekeeping genes GAPDH and actin are shown as loading controls. (B) Flow cytometric analysis of CAL-1 cells after staining with antibodies directed against the cell surface markers BDCA2, CD123, CCR7, CD45RA, CD62L, and CD56 (black lines). Isotype control stainings are shown as gray filled histograms. (C) Surface expression of the costimulatory molecules CD40, CD80, CD86, and HLA-DR was measured by flow cytometry on unstimulated CAL-1 (gray lines) and after 20 hours stimulation with CpG-B (black lines; isotype control stainings are shown as gray filled histograms). (D) CAL-1 cells were stimulated for 4 hours with CpG-B or left unstimulated in medium only, and gene expression levels of CD40, IFN-α and IFN-β1 were measured in stimulated versus unstimulated cells by qPCR. (E) CAL-1 cells were cultured in the presence of CpG-B or medium for 6 hours. Culture supernatants were analyzed for the presence of TNF-α and IL-6 by cytokine bead array analysis. ND indicates not detectable (below detection sensitivity of the assay of 4 pg/mL).
Spi-B is required for leukemic pDC growth and survival in vitro and in vivo
Spi-B is highly expressed in pDCs, but it remains unresolved which target genes are controlled by Spi-B. To gain insight in this, CAL-1 cells were transduced with the lentiviral vector pTRIP expressing either shRNA targeting Spi-B mRNA or the nonexpressed renilla mRNA as a control. The Spi-B shRNAs efficiently reduced the levels of Spi-B mRNA as well as protein (supplemental Figures 2-4).6 High transduction efficiencies (> 95%) were obtained as measured by expression of GFP, which was independently driven from the EF1α promoter in the lentiviral construct. On culture of the transduced cells we noted that the percentage of Spi-B shRNA expressing CAL-1 cells decreased over time unlike cells expressing control shRNA, where the percentage of GFP+ cells remained constant (Figure 2A). Reducing the Spi-B levels in CAL-1 cells led to a significant 1.5-fold reduction in absolute cell numbers compared with control transduced CAL-1 cells 13 days after transduction (Figure 2B). To further examine whether Spi-B is important in cell growth and/or cell survival we sorted GFP+ Spi-B shRNA transduced cells and cultured them in the presence of tritiated thymidine (3H-TdR). Reducing Spi-B expression levels impaired 3H-TdR incorporation in CAL-1 by 2.5-fold after 24 hours (data not shown). Impaired thymidine incorporation was not because of decreased cell proliferation, as shown by equal loss of CellTrace-violet by Spi-B shRNA and control shRNA transduced cells over time (Figure 2C). This prompted us to analyze whether induction of apoptosis could explain reduced 3H-TdR incorporation in Spi-B shRNA transduced CAL-1 cells. Therefore, we sorted transduced GFP+ cells and stained with annexin V and 7-AAD and analyzed percentages of apoptotic cells after 5 days. We observed up to 1.5-fold more annexin V+7-AAD− early apoptotic cells and annexin V+7-AAD+ late apoptotic/necrotic cells in cultures with Spi-B shRNA cells compared with control shRNA transduced cells (Figure 2D), which was consistent when we analyzed this on consecutive days (Figure 2E). Collectively, these results suggest that Spi-B exerts a role in cell survival in the CAL-1 cell line in vitro.
Spi-B is required for CAL-1 cell survival and proliferation. (A) CAL-1 cells were transduced with Spi-B shRNA or control renilla shRNA and cultured for 14 days. Cell growth was determined by flow cytometric analysis to measure the percentages of GFP+ cells every 3 days. (B) Mean values of 3 experiments in which GFP+ cell percentages were determined at 13 days after transduction. Values were normalized to the percentage of GFP+ cells at day 2 after transduction (A; *P = .026). (C) CAL-1 cells transduced with Spi-B shRNA or control renilla shRNA were stained with the CellTrace violet proliferation kit. Mean fluorescence intensity (MFI) was followed for 3 days by flow cytometry. (D) To assay for apoptosis, annexin V–PE and 7-AAD double staining was performed on CAL-1 cell transduced with Spi-B shRNA or control renilla shRNA 5 days after sorting GFP+ cells. Numbers represent percentages of early apopotic cells (annexin V+7-AAD−) and late apoptotic/necrotic cells (annexin V+7-AAD+) in the indicated gates. One representative experiment of 3 is displayed. (E) The percentages of apoptotic cells were measured over time in Spi-B shRNA or control renilla shRNA transduced cells after sorting GFP+ cells. One representative experiment is shown of 2. (F) RAG-2−/−γc−/− immunodeficient mice were injected subcutaneously in the right and left flank with 0.25.106 Cal-1 cells expressing Spi-B shRNAs or renilla shRNAs. Tumor growth is shown as absolute cell numbers measured 20 days after engraftment (*P = .037). Each dot/square represents 1 mouse.
Spi-B is required for CAL-1 cell survival and proliferation. (A) CAL-1 cells were transduced with Spi-B shRNA or control renilla shRNA and cultured for 14 days. Cell growth was determined by flow cytometric analysis to measure the percentages of GFP+ cells every 3 days. (B) Mean values of 3 experiments in which GFP+ cell percentages were determined at 13 days after transduction. Values were normalized to the percentage of GFP+ cells at day 2 after transduction (A; *P = .026). (C) CAL-1 cells transduced with Spi-B shRNA or control renilla shRNA were stained with the CellTrace violet proliferation kit. Mean fluorescence intensity (MFI) was followed for 3 days by flow cytometry. (D) To assay for apoptosis, annexin V–PE and 7-AAD double staining was performed on CAL-1 cell transduced with Spi-B shRNA or control renilla shRNA 5 days after sorting GFP+ cells. Numbers represent percentages of early apopotic cells (annexin V+7-AAD−) and late apoptotic/necrotic cells (annexin V+7-AAD+) in the indicated gates. One representative experiment of 3 is displayed. (E) The percentages of apoptotic cells were measured over time in Spi-B shRNA or control renilla shRNA transduced cells after sorting GFP+ cells. One representative experiment is shown of 2. (F) RAG-2−/−γc−/− immunodeficient mice were injected subcutaneously in the right and left flank with 0.25.106 Cal-1 cells expressing Spi-B shRNAs or renilla shRNAs. Tumor growth is shown as absolute cell numbers measured 20 days after engraftment (*P = .037). Each dot/square represents 1 mouse.
To substantiate the role of Spi-B in cell survival we performed in vivo cell growth experiments using an immune-deficient mouse model in which we monitored CAL-1 cell growth. We inoculated RAG-2−/−γc−/− mice, lacking mouse T, B, and NK lymphocytes,28,29 with sorted GFP+ CAL-1 cells. Simultaneously, mice were injected subcutaneously in the right and left flank with either 0.25 × 106 CAL-1 cells expressing Spi-B shRNA or renilla shRNA, respectively (Figure 2F). Matrigel was used to mimic a solid tumor like structure. Twenty days after inoculation, mice were killed and the tumor tissue was removed and analyzed. By flow cytometry, we confirmed that the tumor cells were still GFP+CD123+CD4+ CAL-1 cells (supplemental Figure 3, data not shown). After counting cell numbers we found that decreased Spi-B levels in CAL-1 cells resulted in significantly reduced subcutaneous growth reaching on average more than 2-fold lower cell numbers than control transduced tumors (Figure 2F). Hardly any GFP+ CAL-1 cells were detected in blood or organs, such as spleen and liver (supplemental Figure 3). Formally we cannot exclude the possibility that CAL-1 cells emigrated from the tumor and migrated to other sites that we did not analyze, or migrated and then died. However, when taken together we conclude that Spi-B controls the growth and survival of this pDC cell line.
Spi-B regulates expression of Bcl2-A1
To elucidate the mechanism by which Spi-B controls cell survival we performed a microarray analysis to identify Spi-B induced target genes. We compared gene expression levels in CAL-1 cells with elevated Spi-B levels (after transduction with pLZRS–Spi-B–IRES-GFP) or decreased Spi-B levels (after transduction with pTRIP–Spi-B–shRNA/EF1α-GFP) and their respective controls using Affymetrix chips, which cover approximately 21 000 known genes of the NCBI database (data not shown). Among the presumptive Spi-B targets identified after bioinformatical analysis, we focused on genes known to be responsible for cell survival. Interestingly, Bcl2-A1 expression correlated with Spi-B levels in CAL-1 cells (data not shown). To validate this microarray result, we measured expression of Bcl2-A1 in sorted GFP+ CAL-1 cells by qPCR 48 hours after transduction with either pLZRS–Spi-B–IRES-GFP, pTRIP–Spi-B–shRNA/EF1α-GFP, or appropriate control constructs. Spi-B overexpression resulted in a 6-fold increase in Bcl2-A1 levels (P < .05), whereas Bcl2-A1 levels were reduced 2.5-fold after knocking down Spi-B expression (P < .01; Figure 3A). To assess potential off-target effects of the Spi-B shRNA, we analyzed expression of several proteins including Bcl-2, Bcl-xL, CALCRL, and IRF-7, and found no significant differences between Spi-B and control shRNAs (supplemental Figure 4). To gain more insight in the regulation of Bcl2-A1 by Spi-B we used an inducible Spi-B–ER fusion construct. Nuclear translocation of Spi-B was induced by this construct in the presence, but not in the absence of 4HT.13 Also we used the protein translation inhibitor cycloheximide (CHX), which is indicative for direct regulation of target genes, in combination with short-term induction of Spi-B with 4HT. Using this approach a 4-fold up-regulation of Bcl2-A1 was observed by QPCR in Spi-B∼ER transduced CAL-1 cells already 4 hours after Spi-B induction (Figure 3B), suggesting that Bcl2-A1 may be a direct target gene of Spi-B. As a positive control we confirmed direct regulation of CD40 expression by Spi-B (Figure 3C).13 Our results support the notion that Spi-B dependent induction of Bcl2-A1 is comparable with CD40 induction under the same conditions. Notably, expression of other antiapoptotic genes, including Bcl-2, Mcl-1, and Bcl-xL, was not up-regulated by overexpression of Spi-B (Figure 3D). Thus, our data provide evidence for a specific role of Spi-B to mediate survival of pDCs through Bcl2-A1 induction.
Expression of Bcl2-A1 is regulated by Spi-B. (A) CAL-1 cells were transduced with vectors expressing Spi-B cDNA, Spi-B shRNAs or appropriate control vectors and sorted for GFP expression after 48 hours. Bcl2-A1 mRNA expression levels were measured by qPCR in 3 independent experiments. Values are normalized to control transduced cells (*P < .05; **P < .01). (B-C) Short term induction of Spi-B using Spi-B∼ER transduced cells incubated with (black bar) or without (white bar) 4HT after pretreatment with cycloheximide to avoid de novo protein synthesis. Bcl2-A1 (B) and CD40 (C) mRNA expression levels were measured by qPCR. Values were normalized to control transduced cells incubated without 4HT. (D) mRNA levels of antiapoptotic genes of the Bcl2 family were measured by QPCR in Spi-B∼ER–transduced CAL-1 cells after 4 hours of incubation in the presence (black bars) or absence (white bars) of 4HT. Values were normalized to transduced cells incubated without 4HT.
Expression of Bcl2-A1 is regulated by Spi-B. (A) CAL-1 cells were transduced with vectors expressing Spi-B cDNA, Spi-B shRNAs or appropriate control vectors and sorted for GFP expression after 48 hours. Bcl2-A1 mRNA expression levels were measured by qPCR in 3 independent experiments. Values are normalized to control transduced cells (*P < .05; **P < .01). (B-C) Short term induction of Spi-B using Spi-B∼ER transduced cells incubated with (black bar) or without (white bar) 4HT after pretreatment with cycloheximide to avoid de novo protein synthesis. Bcl2-A1 (B) and CD40 (C) mRNA expression levels were measured by qPCR. Values were normalized to control transduced cells incubated without 4HT. (D) mRNA levels of antiapoptotic genes of the Bcl2 family were measured by QPCR in Spi-B∼ER–transduced CAL-1 cells after 4 hours of incubation in the presence (black bars) or absence (white bars) of 4HT. Values were normalized to transduced cells incubated without 4HT.
BCL2-A1 is a direct target gene of Spi-B
Our results showing that Bcl2-A1 is regulated by Spi-B prompted us to determine whether Spi-B directly binds the promoter region of BCL2-A1. Therefore, we performed ChIP assays using an anti-ER antibody or unspecific rabbit IgG on lysates derived from Spi-B∼ER transduced CAL-1 cells that were either incubated in medium only or with 4HT for 4 hours. Chromatin abundance was determined by QPCR using different primer sets designed to amplify several regions of the promoter sequence of BCL2-A1 (Figure 4A). We analyzed Spi-B binding to the BCL2-A1 gene locus up to 3.2 kb upstream of the transcriptional start site, and observed direct binding of Spi-B∼ER to the BCL2-A1 promoter on 2 of the 5 regions investigated only when 4HT was added (Figure 4A). The relative quantity of precipitation of these regions with Spi-B was comparable with the relative amount of precipitation of the CD40 promoter sequence, which was previously reported to be bound by Spi-B13 (Figure 4B).
BCL2-A1 is a direct target of Spi-B. (A) Binding of Spi-B to the BCL2-A1 promoter region (3.2 kb) was assessed by ChIP of the Spi-B∼ER fusion protein using an anti-ER antibody or irrelevant IgG control antibody (IgG) in CAL-1 cells incubated with or without 4HT. Pulled down DNA was purified and amplified using 5 different primer sets (PR1-PR5). (B) ChIP analysis of the known binding region of Spi-B to the CD40 promoter region was used as a positive control using the PR+ primer set. One representative ChIP experiment of 4 is depicted. (C) Setup of the dual luciferase assay is shown on top. The full-length region of the BCL2-A1 promoter containing the 2 Spi-B binding sites (black dots) or lacking the 5′ Spi-B binding site (Δ5′) were subcloned into the pGL3-firefly luciferase backbone. Full-length or Δ5′ reporter constructs were cotransfected in 293T cells with vectors expressing the wild-type Spi-B cDNA (wtSpi-B) or mutated cDNAs of Spi-B, either lacking the transactivation domain (ΔTAD) or the Ets domain (ΔEts). Firefly luciferase activity was normalized to renilla reniformis luciferase activity for transfection efficiency. Then, firefly/renilla activity was normalized to control (empty vector), which was set to 1.
BCL2-A1 is a direct target of Spi-B. (A) Binding of Spi-B to the BCL2-A1 promoter region (3.2 kb) was assessed by ChIP of the Spi-B∼ER fusion protein using an anti-ER antibody or irrelevant IgG control antibody (IgG) in CAL-1 cells incubated with or without 4HT. Pulled down DNA was purified and amplified using 5 different primer sets (PR1-PR5). (B) ChIP analysis of the known binding region of Spi-B to the CD40 promoter region was used as a positive control using the PR+ primer set. One representative ChIP experiment of 4 is depicted. (C) Setup of the dual luciferase assay is shown on top. The full-length region of the BCL2-A1 promoter containing the 2 Spi-B binding sites (black dots) or lacking the 5′ Spi-B binding site (Δ5′) were subcloned into the pGL3-firefly luciferase backbone. Full-length or Δ5′ reporter constructs were cotransfected in 293T cells with vectors expressing the wild-type Spi-B cDNA (wtSpi-B) or mutated cDNAs of Spi-B, either lacking the transactivation domain (ΔTAD) or the Ets domain (ΔEts). Firefly luciferase activity was normalized to renilla reniformis luciferase activity for transfection efficiency. Then, firefly/renilla activity was normalized to control (empty vector), which was set to 1.
To test whether binding of Spi-B to the BCL2-A1 promoter is functionally relevant, Luciferase reporter assays were performed. We used a pGL3 vector expressing the firefly luciferase gene controlled by the BCL2-A1 promoter, containing either the 2 putative Spi-B binding sites (full-length), or only the 3′ Spi-B binding site (Δ5′; Figure 4C). Cotransfection of the full-length reporter construct together with a Spi-B expression vector in 293T cells resulted in 4-fold induction of luciferase activity (Figure 4D). Deletion of the 5′ Spi-B binding site (Δ5′) resulted in 2-fold lower luciferase induction by Spi-B compared with the full-length BCL2-A1 promoter reporter construct. We also analyzed 2 natural splicevariants of Spi-B, ΔEts–Spi-B, lacking the DNA binding ETS domain, and ΔTAD–Spi-B, lacking the transactivation domain. Notably, the BCL2-A1 promoter reporter construct was completely unresponsive to these Spi-B mutants. Introduction of the wild type Spi-B together with ΔTAD–Spi-B rescued luciferase activity. Activation of the BCL2-A1 promoter reporter by Spi-B was of similar magnitude as activation by its homologue PU.1, which was previously shown to transactivate this promoter.30 These observations, together with the ChIP results and lower Bcl2-A1 expression after reducing Spi-B levels, strongly suggest that Spi-B directly controls expression of the BCL2-A1 gene.
Bcl2-A1 is required for in vitro pDC development
Next, we aimed to decipher the physiologic relevance of Bcl2-A1 regulation by Spi-B during human pDC development. We designed 3 different shRNAs to target Bcl2-A1 mRNA, of which shRNA1 and shRNA3 resulted in 40% knockdown in mRNA levels when transduced in Bcl2-A1 expressing CAL-1 cells (supplemental Figure 5). We used Bcl2-A1 shRNA3 in addition to renilla control shRNA to transduce FL CD34+CD38− HPCs and performed in vitro human pDC differentiation assays as reported previously.9 We observed a significant decrease of 25% in absolute GFP+CD123+BDCAs-2+ pDC cell numbers when overexpressing Bcl2-A1 shRNAs in HPCs compared with control shRNAs after 7 days of culture (Figure 5, n = 15, P = .003; supplemental Table 2). It is interesting that this effect was specific for pDC development as other cell types (non-pDCs) that differentiated in this culture (including monocytic cells, not shown) were not significantly affected. Taken together, these data show that development of pDCs from CD34+ HPCs depends on Bcl2-A1 expression. In combination with our previous observation that Spi-B is a master regulator of pDC development,6 these results lead us to propose that Spi-B mediates the survival of pDCs and possibly its progenitor cells by preventing apoptosis through induction of the antiapoptotic gene BCL2-A1.
Bcl2-A1 is required for pDC differentiation in vitro. CD34+CD38− HPCs from human FL were transduced with vectors expressing Bcl2-A1 shRNAs or renilla control shRNAs, and cocultured with OP9 stromal cells with IL-7 and Flt3L for 7 days. Flow cytometric analysis was performed to identify BDCA2+CD123+GFP+ in vitro generated pDCs or GFP+BDCA-2− non-pDCs. Shown are the absolute cell numbers normalized to the control culture of 15 donors (*P = .012, ***P = .003). Calculations were done as described in “Methods.”
Bcl2-A1 is required for pDC differentiation in vitro. CD34+CD38− HPCs from human FL were transduced with vectors expressing Bcl2-A1 shRNAs or renilla control shRNAs, and cocultured with OP9 stromal cells with IL-7 and Flt3L for 7 days. Flow cytometric analysis was performed to identify BDCA2+CD123+GFP+ in vitro generated pDCs or GFP+BDCA-2− non-pDCs. Shown are the absolute cell numbers normalized to the control culture of 15 donors (*P = .012, ***P = .003). Calculations were done as described in “Methods.”
Discussion
Here, we identified Bcl2-A1 as a direct transcriptional target of Spi-B in pDCs. As predicted by micro-array analysis, overexpression of Spi-B in a pDC cell line up-regulated Bcl2-A1. In contrast, Spi-B knockdown reduced BCL2-A1 transcription, which had direct consequences for cell survival both in vitro and in vivo. Direct binding of Spi-B to the Bcl2-A1 promoter was confirmed by ChIP assays. More importantly, in vitro human pDC differentiation from CD34+ HPCs, which crucially depends on Spi-B,6 was impaired when inhibiting expression of Bcl2-A1. Taken together, these findings support a role for Spi-B as regulator of pDC survival.
Spi-B is involved at distinct steps during normal hematopoiesis, including differentiation of pDCs,9 plasma cells,13 and maintenance of follicular B cells.31 It is incompletely understood what the critical target genes are in these differentiation steps. Several targets were identified previously including CD40,13 Grap2,32 P2Y10,33 c-rel,34 c-fes/c-fps,7 and Btk.35 Most of these targets were identified in mice, except CD40, which is also in human cells controlled by Spi-B.13 We were unable to confirm the other reported genes as candidate targets of Spi-B in human cells, either using microarray analysis or QPCR (data not shown). Although this may be explained by mouse versus human species differences, it may also reflect cellular differences as we used a human pDC cell line, whereas others used microglial cells, macrophages, or B cells. Except for c-rel and Btk13,36 none of the other target genes mentioned are known to be expressed in human pDCs. It is interesting to understand how expression of Btk and c-rel is regulated in pDCs.
Expression of most reported target genes relied on the transactivating potential of Spi-B critically depending on both the ETS and TAD domains.13 We recently observed that Spi-B has repressor activity as well. Expression of the plasma cell genes PRDM1 and XBP-1 in human B cells was impaired by direct binding of Spi-B to their promoters.13 This relied on the ETS domain, but not the TAD domain, suggesting that DNA binding is requisite for repressor activity. A similar dual role was reported for PU.1 (reviewed in Marecki and Fenton37 ), which is most homologous to Spi-B.8 Whereas the differential role of Spi-B as a transcriptional repressor or activator has not been characterized in much detail, for PU.1 it was reported that putative acetylation motifs within the ETS domain were crucial for its repressor activity.38 These motifs may mediate the interplay between PU.1 and other proteins, including B-cell lymphoma 6 (BCL-6) or IRF-8, which control the repressor or activator activity of PU.1, respectively.37 Interestingly, Spi-B also contains putative acetylation motifs in its ETS domain (data not shown). Moreover, Spi-B interacts with BCL-639 and IRF members,40 suggesting that the repressor or activator function of Spi-B may be regulated similarly to PU.1. Whether regulation of cell survival induced by Spi-B involves such heterodimeric protein complexes is unknown. But as Spi-B binds the promoter of Bcl2-A1 in pDCs and promotes Bcl2-A1 expression, it seems probable that Spi-B induced cell survival depends on its transactivation properties. In accordance with this, we observed that not only the ETS domain, but also the TAD domain of Spi-B was essential to induce luciferase activity from the Bcl2-A1 promoter construct.
Bcl2-A1 and Bcl-xL are also induced after activation of NF-κB.24,41 In line with this, we observed that activation of freshly isolated pDCs through engagement of TLR-9 with CpG-ODN, which elicits strong activation of NF-κB, induced Bcl2-A1 expression, as well as other antiapoptotic proteins, such as Mcl-1, Bcl-xL, and Bcl-2 (data not shown). In mice, the NF-κB subunit c-rel was reported as direct target of Spi-B,34 although we were unable to confirm this in human cells. More importantly, it is unlikely that Bcl2-A1 induction in our study is the result of Spi-B–induced c-rel mRNA expression, because Bcl2-A1 induction was observed in the presence of cycloheximide blocking de novo protein synthesis (Figure 3B). This, together with our finding that Spi-B binds the BCL2-A1 promoter suggests direct regulation of Bcl2-A1 by Spi-B, in the absence of TLR triggering and NF-κB activation.
In vitro, pDC rapidly die by apoptosis induction.14 Cell survival can be induced by addition of cytokines (GM-CSF, IL-3)14 or TLR agonists,36 mediating up-regulation of antiapoptotic genes including BCL-XL, BCL-2, BIRC3, cFLAR,42 and BCL2-A1.43 In vivo, mouse pDCs are long lived cells.14 Consistent with this, the lifespan of pDCs in healthy, noninfected animals was 5-times longer than the lifespan of spleen CD8+ DCs.44 It is tempting to speculate that Spi-B controls pDCs homeostasis in vivo by up-regulation of Bcl2-A1. Whereas the number of pDCs in bcl2-a1−/− mice is undetermined, neutrophil numbers in these mice were reduced because of enhanced apoptosis.45 Neutrophils do not express Spi-B, but do express high levels of PU.1.46 Because PU.1 and Spi-B are highly homologues,8 and PU.1 binds the Bcl2-A1 promoter,30 this may explain the loss of neutrophils in bcl2-a1−/− mice. As pDCs lack PU.1 expression,6 but do express high levels of Spi-B,9 we propose that apoptosis inhibition in pDC in vivo may be controlled by Spi-B induced expression of Bcl2-A1.
Impaired pDC differentiation when reducing Bcl2-A1 levels by shRNAs could partially be rescued by overexpression of Spi-B (data not shown). However, it is noteworthy that Bcl2-A1 overexpression failed to rescue pDC development when HPCs were transduced with Spi-B shRNA (data not shown). This was not entirely unexpected, as it highlights the role of Spi-B as master regulator in pDC development. Previously, we showed that overexpression of Spi-B in HPCs down-regulated expression of inhibitor-of-DNA binding (Id)2 thereby allowing E2-2 activity to promote pDC development.11 Hence, no single factor downstream of Spi-B may fully recapitulate the effects of Spi-B itself.
The leukemic pDC cell line CAL-1 expresses Spi-B, and depended on Spi-B for its survival as overexpression of Spi-B shRNAs was toxic for the cells. Our findings support the notion that Spi-B has an oncogenic role in pDC leukemic cells by inducing Bcl2-A1. Aberrant expression of Spi-B has also been implicated in tumorigenesis in human B cells. Spi-B transcripts were detected in several B-cell derived malignancies, including multiple myeloma cells and B-chronic lymphocytic leukemia (B-CLL).47 In addition, high levels of Spi-B were detected in activated B cell-like diffuse large B-cell lymphoma (ABC-DLBCL) either because of translocation of SPI-B to the Ig heavy-chain locus or to amplification of a chromosomal segment including the Spi-B locus.48 Interestingly, reducing Spi-B expression levels in an ABC-DLBCL cell line prevented cell survival as well.49 Although the underlying mechanism was not investigated, it is tempting to speculate that Spi-B controls Bcl2-A1 expression in ABC-DLBCL cells supporting its poor clinical outcome. Taken together, this illustrates that Spi-B may be an attractive candidate gene for drug targeting in diverse types of lymphomas. Hence, expanding our knowledge on Spi-B and its target genes not only contributes to our general understanding of normal lymphocyte development, but in addition will be central to provide detailed insight in its role during oncogenesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Berend Hooibrink and Toni van Capel for maintaining the FACS facility. They acknowledge Prof Hazekamp and staff from the Leids Universitair Medisch Centrum (Leiden, The Netherlands) for providing human thymus tissue, the Bloemenhove clinic (Heemstede, The Netherlands) and staff for providing fetal tissues, and Dr Kees Weijer and Arie Voordouw for collecting fetal tissues. They thank Dr R. Schotte and Dr N. Legrand for critically reading the paper.
J.J.K. is supported through a personal VIDI grant to B.B. (Dutch Science Foundation, no. 917.66.310). M.B. is supported through a personal VENI grant (Dutch Science Foundation) to M.C.W.
Authorship
Contribution: J.J.K. designed research, performed experiments, analyzed data, and wrote the paper; M.B. and H.S. designed research, performed experiments, analyzed data; M.L., M.N., and R.G. performed experiments and analyzed data; S.K. and T.M. contributed essential reagent; D.A. and M.C.W. analyzed data; and B.B. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bianca Blom, Academic Medical Center, University of Amsterdam, Meibergdreef 15, Amsterdam, The Netherlands; e-mail: b.blom@amc.uva.nl.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal