In response to microenvironmental signals, macrophages undergo different activation, including the “classic” proinflammatory phenotype (also called M1), the “alternative” activation induced by the IL-4/IL-13 trigger, and the related but distinct heterogeneous M2 polarization associated with the anti-inflammatory profile. The latter is induced by several stimuli, including IL-10 and TGF-β. Macrophage-polarized activation has profound effects on immune and inflammatory responses and in tumor biology, but information on the underlying molecular pathways is scarce. In the present study, we report that alternative polarization of macrophages requires the transcription factor c-MYC. In macrophages, IL-4 and different stimuli sustaining M2-like polarization induce c-MYC expression and its translocation to the nucleus. c-MYC controls the induction of a subset (45%) of genes associated with alternative activation. ChIP assays indicate that c-MYC directly regulates some genes associated with alternative activation, including SCARB1, ALOX15, and MRC1, whereas others, including CD209, are indirectly regulated by c-MYC. c-MYC up-regulates the IL-4 signaling mediators signal transducer and activator of transcription-6 and peroxisome proliferator–activated receptorγ, is also expressed in tumor-associated macrophages, and its inhibition blocks the expression of protumoral genes including VEGF, MMP9, HIF-1α, and TGF-β. We conclude that c-MYC is a key player in alternative macrophage activation, and is therefore a potential therapeutic target in pathologies related to these cells, including tumors.
Introduction
Macrophages are specialized phagocytic cells involved in multiple processes, both in homeostatic conditions and during the immune response after tissue damage or exposure to a pathogen. Macrophages are characterized by a striking heterogeneity, which can be partially ascribed to their origin by self-renewal of resident postmitotic cells and by monocyte subsets recruited and differentiated locally.1,–3 A second element shaping macrophage heterogeneity is the microenvironment, both under homeostatic conditions, with the hosting tissue profoundly influencing macrophage differentiation, and in the context of an inflammatory or immune response, which generates a wide range of polarized activation states.3,,–6 Activation with IFN-γ, alone or in combination with pathogen-derived signals such as lipopolysaccharide (LPS), leads to classically activated macrophages, also referred to as M1 cells, which develop proinflammatory type 1 immune responses. Macrophage exposure to other immune signals results in profoundly different functional phenotypes. These include “alternatively activated” macrophages caused by IL-4/IL-13 stimulation, which are associated with type 2 immune responses and a spectrum of functional phenotypes related to anti-inflammatory, angiogenic, and tissue-repair properties induced in macrophages by stimuli including TGF-β, immune complexes, glucocorticoids, and IL-10.3,4,6,7 Furthermore, in several tumor types, tumor-associated macrophages (TAMs) resemble alternatively activated macrophages in several respects,8,9 although the existence of TAM subsets with distinct functional properties has been reported recently.10,11 At present, our understanding of the molecular mechanisms sustaining macrophage-polarized activation in response to microenvironment-derived signals is fragmentary and incomplete. Regarding alternative macrophages, it is known that peroxisome proliferator–activated receptorγ (PPARγ) skews monocytes toward an M2 phenotype12 and down-regulates inflammatory pathways in mature macrophages.13 Alternative macrophage activation is also favored by transcriptional regulators interfering with the NF-κB pathway.14 More recently, a pathway involving the IFN regulatory factor-4 (IRF4) and the histone demethylase jumonji domain containing-3 (JMJD3) has been shown to regulate a restricted subset of M2 markers directly.15,16 Nevertheless, the molecular entities involved in the global rearrangement of the transcriptional profile occurring during alternative macrophage activation are still largely unknown.
c-MYC is a protooncogene with a deregulated expression that is associated with the development of tumors in mice and humans; for this reason, its role in tumor cell biology has been investigated extensively.17 This transcription factor is also involved in several processes in nontransformed cells, including cell growth and apoptosis/survival.18 Resting cells normally express low levels of c-MYC, which as an immediate early-response gene is dramatically increased by exposure to growth factors.19,–21 c-MYC then acts as a transcription factor by interacting with the protein MAX.22 The c-MYC/MAX heterodimer binds with high affinity to the CACGTG E-box sequence in the promoter region of target genes and induces their expression.22,23 In addition, the c-MYC/MAX complex can suppress the expression of certain genes24 in a process that is mostly related to the protumoral activity of c-MYC.25 More recently, a key role of this transcription factor in hematopoietic stem-cell function and survival and in lymphoid compartment homeostasis has also been reported.26,,–29
In the present study, we report that c-MYC is induced in human macrophages activated by IL-4– and M2-like stimuli, and provide evidence for its involvement in the induction of a large set of genes during alternative macrophage activation, either by direct interaction or indirectly through induction of signal transducer and activator of transcription-6 (STAT6) and PPARγ. We also report that c-MYC is expressed in TAMs, where it controls the expression of protumoral genes.
Methods
Reagents
Recombinant human and mouse cytokines were obtained from PeproTech. LPS from Escherichia coli (serotype 055:B5), cycloheximide, and the PPARγ antagonist GW9662 were obtained from Sigma-Aldrich. Accutase, the c-MYC inhibitor 10058-F4, and the JAK2/3 inhibitor AG490 were purchased from Calbiochem. The anti–c-MYC polyclonal antiserum N-262 was purchased from Santa Cruz Biotechnology. Other Abs were purchased from Serotec unless otherwise specified.
Cell cultures
The human tumor cell lines PANC-1 (from pancreatic tumor), SW480 (from colon tumor), and MDA–MB-231 (from breast tumor) were from the ATCC. Conditioned media were obtained and used as described previously.30 Human monocytes were obtained from healthy donor buffy coats by 2-step gradient centrifugation using Ficoll (Biochrom) and Percoll (Amersham). Nonadherent cells were discarded, and the purified monocytes were incubated for 7 days in RPMI 1640 medium (Biochrom) supplemented with 10% FCS (HyClone) and 100 ng/mL of M-CSF to obtain resting macrophages. Macrophage polarization was obtained by removing the culture medium and culturing cells in RPMI 1640 medium supplemented with 10% FCS and 20 ng/mL of IFNγ plus 100 ng/mL of LPS (M1 polarization) or 20 ng/mL of IL-4 (alternative activation) for 24 hours.7 Where indicated, chemical inhibitors or their corresponding vehicles were added during macrophage polarization. Shifts in polarizing conditions were obtained by substituting medium conditions and culturing cells for an additional 24 hours. Cell-cycle analysis was performed using the PI/RNase Staining Kit (BD Pharmingen) and evaluated by flow cytometry (FACSCanto II; BD Biosciences). The apoptosis index was analyzed using the Cell Death Detection Kit (Roche) and evaluated by flow cytometry (FACSCanto II; BD Biosciences). Peritoneal macrophages were harvested by rinsing the peritoneal cavity twice with 5-mL washes of PBS, and plated in culture in RPMI 1640 medium supplemented with 10% FCS. After 8 hours, nonadherent cells were removed. More than 90% of adherent cells were macrophages.
Western blot analysis
After removing the medium, cells were washed in PBS and lysed in ice-cold lysis buffer (2% Triton X-100, 10mM Tris-HCl, pH 8.0, 150mM NaCl, 2mM NaN3, and 2mM EDTA) containing protease inhibitors (Roche) for 45 minutes at 4°C. Lysates were harvested and centrifuged at 13 400g to eliminate nuclei. The protein concentration was determined using the bicinchoninic acid assay (Pierce), and 30 μg of protein was electrophoresed in a 10% SDS-PAGE under reducing conditions and transferred to nitrocellulose using standard procedures.
Microarray analysis
The transcriptional profile was evaluated in 3 independent preparations of resting and IL-4–treated macrophages in the presence of the c-Myc chemical inhibitor 10058-F4 or its vehicle. Each preparation was analyzed using the Human Genome HuGene 1.0 ST (Affymetrix) and Partek Version 6.3 software. Genes with an induction of ≥ 2-fold and P ≤ .05 were considered to be in M2 (as opposed to M0) and were considered M2 markers. M2 markers were considered-MYC dependent when P ≤ .05 comparing IL-4 with IL-4 plus c-MYC inhibition conditions. A heat-map analysis of this set of genes was made using the Pearson correlation as a similarity measure, and TIGR MultiExperiment Viewer 4.7 software (Dana-Farber Cancer Institute).31 The percentage of inhibition was calculated as 100 − [(Y × 100) / X], where X = mean FC M2 − mean FC M0 and Y = mean FC M2 plus inhibitor − mean FC M0. The microarray data are available in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/gds) under accession number GSE32164.
Bioinformatic analysis of transcription binding sites
Consensus sequences for c-MYC and STAT6 within the target genes' putative promoter region (ie, the 2 kb upstream from the transcription start site) were identified using the Human Genome browser (http://genome.ucsc.edu/) and the ALGGEN program (http://alggen.lsi.upc.es/).
Gene-expression analysis by real-time quantitative PCR (Q-PCR)
Total RNA was isolated from the cell cultures with TRIzol reagent (Invitrogen) according to the manufacturer's instructions, and was quantified by its absorption at 260 nm. First-strand cDNA was synthesized from 2 μg of total RNA in 25 μL of reaction mixture using the High-Capacity cDNA Achieve kit (Applied Biosystems). Gene-specific primers for Q-PCR were designed using Autoprime 1.0 software (www.autoprime.de) and are reported in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Three replicates per each experimental point were performed, and differences were assessed with a 2-tailed Student t test. Results were normalized using the housekeeping GAPDH and the ΔΔ cycle threshold method and are expressed as the relative change (-fold) of the stimulated group over the control group, which was used as a calibrator.
Lentiviral plasmids and macrophage infection
HEK-293T cells were transiently transfected with pLKO.1-puro or pLKO.1-puro-shRNA for c-MYC plus Δ8.74 and VsVg envelopes to obtain lentiviral particles using Lipofectamine 2000 (Invitrogen). Culture supernatants were collected 72 hours after transfection and were used to infect monocyte-derived macrophages. Lentivirus-infected macrophages were monitored for the expression of green fluorescent protein, and the suppression of c-MYC expression in c-MYC/shRNA–infected macrophages was confirmed by Western blot analysis (supplemental Figure 1).
Generation of c-Myc–fl/c-Myc–ERKI mice and estrogen-dependent c-Myc macrophages
Macrophages were derived from c-Myc–fl/c-Myc–ERKI mice, which were generated by crossing c-Myc–floxed mice, in which Cre recombinase inactivates the c-Myc gene via deletion of exons 2 and 332 (kindly provided by Frederick Alt and Moreno De Alboran, Howard Hughes Medical Institute and Children's, Harvard Medical School, Boston, MA), with c-Myc–ERKI mice (kindly provided by Julia Prescott, Department of Pathology, Helen Diller Family Comprehensive Cancer Center, University of California, San Francisco, CA), in which the ligand-binding domain of the mutant 4-hydroxytamoxifen-responsive mutant estrogen receptor has been cloned in-frame at the C-terminus of the c-Myc gene (c-Myc–ERTAM). In the c-Myc–ERKI mouse, the expression of the fusion gene is still controlled by the endogenous c-Myc promoter, but the switchable form ofthe c-Myc protein is functionally active only after exposure to 4-hydroxytamoxifen (4-HT).33 In macrophages derived from c-Myc–fl/c-Myc–ERKI mice, c-Myc expression is controlled by the c-Myc endogenous promoter, but after recombinase-mediated deletion of the c-Myc–floxed allele, c-Myc activity is entirely dependent on exposure to the exogenous agonist 4-HT.
Mice were maintained and treated in accordance with protocols approved by the Institutional Animal Care and Use Committee of the University of California, San Francisco. Macrophages were isolated from the peritoneal cavity of adult c-Myc–fl/c-Myc–ERKI mice and placed in culture in RPMI medium containing 10% FCS. The c-Myc–floxed allele was deleted by incubating macrophages for 72 hours with adeno-cre viral particles (purchased from Gene Transfer Vector Core at the University of Iowa: http://www.uiowa.edu/∼gene) at 3.6 PFU viral particles per 106 cells. Macrophages were then treated for 24 hours with IL-4 (20 ng/mL) and/or 4-HT (100nM). For each condition, macrophages were derived from 3 different animals.
ChIP assay
In brief, 4 × 106 resting macrophages were stimulated for 24 hours with IL-4 (20 ng/mL) or tumoral-conditioned medium (1:3 diluted in RPMI 1640) and fixed by supplementing the medium with formaldehyde (Calbiochem) to a final concentration of 1%. After 10 minutes, ice-cold PBS was added, plates were transferred on ice and washed extensively with PBS, and cells were collected. After centrifugation, cells were lysed for 5 minutes in L1 buffer (50mM Tris, pH 8.0, 2mM EDTA, 0.1% NP-40, and 10% glycerol) supplemented with protease inhibitors. Nuclei were pelleted at 1880g in a microfuge and resuspended in L2 buffer (50mM Tris, pH 8.0, 1% SDS, and 5mM EDTA). Chromatin was sheared by sonication (5 × 10 seconds at 1/5 of the maximum potency in a Sonics Vibracell VC13 equipped with a 3-mm tip), centrifuged to pellet debris, and diluted 10× in dilution buffer (50mM Tris, pH 8.0, 0.5% NP-40, 0.2M NaCl, and 0.5mM EDTA). Extracts were precleared for 2 hours with 80 μL of a 50% suspension of salmon-sperm DNA-saturated protein A. Immunoprecipitation was performed at 4°C overnight with 2 μg of polyclonal anti–human C-MYC or IgG Ab (Santa Cruz Biotechnology). Immune complexes were collected with salmon-sperm DNA-saturated protein A, washed 3 times (5 minutes each) with high-salt buffer (washing buffer: 20mM Tris, pH 8.0, 0.1% SDS, 1% NP-40, 2mM EDTA, and 500mM NaCl), 2 times with a 0.5M LiCl buffer, and 3 times with a low-salt buffer (Tris-EDTA). Immune complexes were extracted in Tris-EDTA containing 2% SDS and protein; DNA cross-links were reverted by heating at 65°C for 6 hours. After tissue digestion with proteinase K (100 μg) for 2 hours at 50°C, DNA was extracted by phenol/chloroform and ethanol precipitated. Approximately 1/20 of the immunoprecipitated DNA was used in each PCR. Gene-specific primers were designed using Autoprime 1.0 software (www.autoprime.de) and are listed in supplemental Table 2. Three replicates for each experimental point were performed. Results were normalized using the internal control IgG and the ΔΔ cycle threshold method and are expressed as the relative change (-fold) of the stimulated group over the control group, which was used as a calibrator.
Immunohistochemistry and confocal microscopy analysis
To determine the nuclear localization of c-MYC, human macrophages were stimulated for 24 hours with 20 ng/mL of IL-4, stained with a rabbit polyclonal anti–c-MYC antiserum, and analyzed with a laser scanning confocal microscope (FluoView FV1000; Olympus). Images were acquired with an oil-immersion objective (40×, 1.3 NA Plan-Apochromat; Olympus). The mean of the fluorescence intensity is expressed as the relative change (-fold) of the stimulated group over the control group, which was used as a calibrator. c-MYC expression in tumor-associated macrophages was investigated by double immunohistochemistry. Frozen sections of normal and neoplastic colon were cut and mounted on SuperFrost slides (Bio-Optica). Slides were fixed in chloroform/acetone for 3 minutes and air-dried. Anti–human CD68 monoclonal Ab (clone PGM1; Dako) at a 1:1000 dilution was used for the first reaction, followed by anti–human c-MYC polyclonal Ab (Santa Cruz) at a 1:100 dilution for the second reaction. A specific biotin-free detection system (MACH2 double-stain polymer kit for mouse and rabbit primary Abs; Biocare Medical) conjugated with alkaline phosphatase and peroxidase was chosen: the first reaction was developed using warp as chromogen (red stained; Biocare Medical) and the second reaction was developed using diaminobenzidine (brown stained; Biocare Medical). Nuclei were counterstained with hematoxylin.
Statistical analysis
All data are presented as means ± SD and are based on experiments performed at least in triplicate. Statistical significance was calculated with the Student t test and Prism 4.03 software (GraphPad).
Results
c-MYC expression in M2 macrophages
Human monocytes and resting macrophages expressed low levels of the c-MYC transcript. Sustained induction of c-MYC was observed after exposure to IL-4 but not under classic conditions (Figure 1A). Western blot analysis confirmed higher expression of the c-MYC protein in M2 cells compared with unstimulated and M1 macrophages (Figure 1B), and confocal microscopy demonstrated an accumulation of c-MYC within the nucleus of macrophages in response to IL-4 exposure compared with resting macrophages (Figure 1C). Finally, Q-PCR analysis demonstrated that c-MYC is also induced in human macrophages in response to several stimuli, thus inducing M2 macrophage activation (Figure 1D). In addition, Q-PCR showed that c-MYC induction is reverted when polarized cells are shifted to M1-polarizing conditions (Figure 1E), indicating that the c-MYC transcription factor is part of the specific signature of alternative macrophage activation.
c-MYC expression in M2 macrophages. (A) Expression levels of the c-MYC transcript in human resting macrophages (○; n = 6 independent donors) and after exposure to 20 ng/mL of IFN-γ plus 10 ng/mL of LPS (□; n = 6 independent donors) or 20 ng/mL of IL-4 (■; n = 6 independent donors) at different time points. (B) Expression levels of c-Myc at protein level in human resting macrophages (M0) and after 48 hours of exposure to 20 ng/mL of IFN-γ plus 10 ng/mL LPS (M1) or 20 ng/mL of IL-4 (M2). Results from 1 experiment representative of 3 performed are shown. (C) Immunofluorescence analysis revealing c-MYC translocation to cell nucleus 48 hours after exposure to 20 ng/mL of IL-4 (inset shows 1 experiment representative of 3 performed, shown as means ± SD). (D) Expression levels of the c-MYC transcript in monocytes (Mono), resting macrophages (M0), and macrophages stimulated for 24 hours with 20 ng/mL of IFN-γ plus 10 ng/mL of LPS (M1), 20 ng/mL of IL-4, 20 ng/mL of IL-13, 20 ng/mL of IL-10, 20 ng/mL of TGF-β, 20 ng/mL of dexamethasone (GC). Results are shown as means ± SD of n = 6 independent experiments. (E) c-MYC induction after exposure to 20 ng/mL of IL-4 is reverted when cells are shifted to M1-polarizing conditions (20 ng/mL of IFN-γ plus 10 ng/mL of LPS) for an additional 24 hours.
c-MYC expression in M2 macrophages. (A) Expression levels of the c-MYC transcript in human resting macrophages (○; n = 6 independent donors) and after exposure to 20 ng/mL of IFN-γ plus 10 ng/mL of LPS (□; n = 6 independent donors) or 20 ng/mL of IL-4 (■; n = 6 independent donors) at different time points. (B) Expression levels of c-Myc at protein level in human resting macrophages (M0) and after 48 hours of exposure to 20 ng/mL of IFN-γ plus 10 ng/mL LPS (M1) or 20 ng/mL of IL-4 (M2). Results from 1 experiment representative of 3 performed are shown. (C) Immunofluorescence analysis revealing c-MYC translocation to cell nucleus 48 hours after exposure to 20 ng/mL of IL-4 (inset shows 1 experiment representative of 3 performed, shown as means ± SD). (D) Expression levels of the c-MYC transcript in monocytes (Mono), resting macrophages (M0), and macrophages stimulated for 24 hours with 20 ng/mL of IFN-γ plus 10 ng/mL of LPS (M1), 20 ng/mL of IL-4, 20 ng/mL of IL-13, 20 ng/mL of IL-10, 20 ng/mL of TGF-β, 20 ng/mL of dexamethasone (GC). Results are shown as means ± SD of n = 6 independent experiments. (E) c-MYC induction after exposure to 20 ng/mL of IL-4 is reverted when cells are shifted to M1-polarizing conditions (20 ng/mL of IFN-γ plus 10 ng/mL of LPS) for an additional 24 hours.
c-Myc is involved in the expression of a distinct subset of alternative activation genes
The functional relevance of c-MYC in alternative macrophage activation was investigated by evaluating the effect of 10058-F4, a c-MYC inhibitor blocking its dimerization with MAX,34 on the transcriptional profile induced on human macrophages by IL-4. In agreement with previous results,2,–4,7,8,35,36 microarray analysis showed that IL-4 induced a restricted set of genes in human macrophages (204 genes; 0.7% of the transcriptome), 45.6% of which were significantly affected by pretreatment with 10058-F4. Therefore, microarray results indicate that alternative activation genes can be classified as c-MYC dependent (Figure 2) or c-MYC independent (supplemental Figure 1) according to the role of the c-MYC transcription factor in their induction during alternative macrophage activation.
c-MYC–dependent genes in M2 macrophages. Microarray analysis was conducted on 3 independent preparations of resting macrophages (M0) and macrophages exposed to 20 ng/mL of IL-4 for 24 hours in the presence or absence of the c-MYC inhibitor 10058-F4 (60μM; M2 inh and M2, respectively). Hierarchical clustering of genes induced by IL-4 (fold of induction ≥ 2 and P ≤ .05 on a 2-tailed paired Student t test comparing M2 vs M0) with induction significantly affected by c-MYC inhibition is shown (P ≤ .05 on a 2-tailed paired Student t test comparing M2 inh and M2). Genes not significantly affected by c-MYC inhibition are shown in supplemental Figure 3. Columns report gene names in the HUGO nomenclature, the -fold of increase of genes in M2 versus M0 in the absence or presence of c-MYC inhibitor (M2/M0 ratio and M2 inh/M0 ratio, respectively), and the percentage and statistical significance of inhibition of gene induction in the presence of c-MYC inhibition.
c-MYC–dependent genes in M2 macrophages. Microarray analysis was conducted on 3 independent preparations of resting macrophages (M0) and macrophages exposed to 20 ng/mL of IL-4 for 24 hours in the presence or absence of the c-MYC inhibitor 10058-F4 (60μM; M2 inh and M2, respectively). Hierarchical clustering of genes induced by IL-4 (fold of induction ≥ 2 and P ≤ .05 on a 2-tailed paired Student t test comparing M2 vs M0) with induction significantly affected by c-MYC inhibition is shown (P ≤ .05 on a 2-tailed paired Student t test comparing M2 inh and M2). Genes not significantly affected by c-MYC inhibition are shown in supplemental Figure 3. Columns report gene names in the HUGO nomenclature, the -fold of increase of genes in M2 versus M0 in the absence or presence of c-MYC inhibitor (M2/M0 ratio and M2 inh/M0 ratio, respectively), and the percentage and statistical significance of inhibition of gene induction in the presence of c-MYC inhibition.
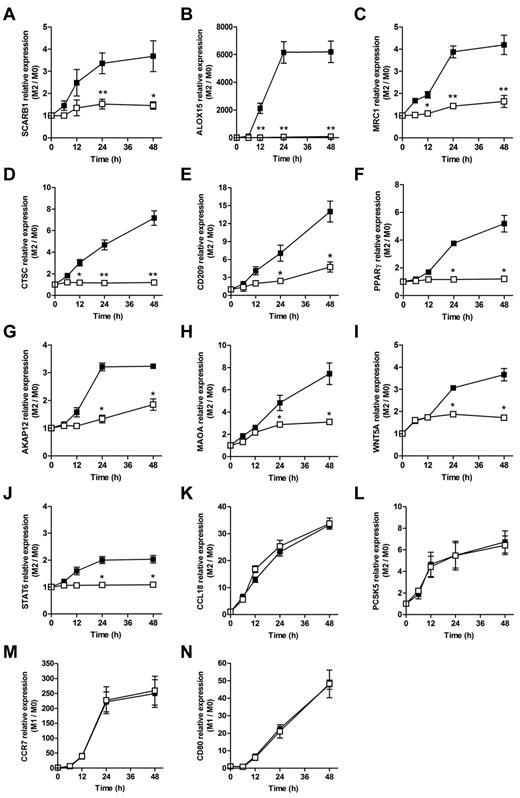
In agreement with microarray data, in the Q-PCR experiments, 10058-F4 showed an inhibitory effect on the IL-4–dependent induction of the scavenger receptor class B (SCARB1; Figure 3A), the lipid-peroxiding enzyme 12/15-lipoxygenase (ALOX15, Figure 3B), the mannose receptor C type 1 (MRC1; Figure 3C), cathepsin C (CTSC; Figure 3D), CD209 (also known as DC-SIGN; Figure 3E), PPARγ (Figure 3F), the A-kinase anchor protein 12 (AKAP12; Figure 3G), the monoamine oxidase A (MAOA; Figure 3H), and the wingless-type MMTV integration site family member 5A (WNT5A; Figure 3I). Interestingly, c-MYC was also involved in the small but reproducible induction by IL-4 of its signal transducer STAT6 (Figure 3J), as demonstrated previously in other cell types.37 Conversely, the IL-4–dependent induction of the CC chemokine ligand 18 (CCL18; Figure 3K) and of the proprotein convertase subtilisin/kexin 5 (PCSK5; Figure 3L) were not affected by c-MYC blockade. c-MYC inhibition had no effect on the induction of gene expression by IFNγ plus LPS stimulation, including CD80 (Figure 3M) and CCR7 (Figure 3N).
c-MYC–dependent IL-4–inducible genes. Expression levels relative to resting macrophages (M0) in macrophages exposed to 20 ng/mL of IL-4, in the presence of the c-MYC inhibitor 10058-F4 (□; 60μM) or its vehicle (■), of SCARB1, ALOX15, MRC1, CTSC, CD209, PPARγ, AKAP12, MAOA, WNT5A, STAT6, CCL18, and PCSK5 (panels A through L, respectively). Expression levels relative to resting macrophages (M0) in macrophages exposed to 20 ng/mL of IFN-γ plus 10 ng/mL of LPS in the presence (□) or absence (■) of the c-MYC inhibitor 10058-F4 (60μM) of CCR7 and CD80 (panels M and N, respectively). Results are shown as means ± SD of n = 5 independent experiments.
c-MYC–dependent IL-4–inducible genes. Expression levels relative to resting macrophages (M0) in macrophages exposed to 20 ng/mL of IL-4, in the presence of the c-MYC inhibitor 10058-F4 (□; 60μM) or its vehicle (■), of SCARB1, ALOX15, MRC1, CTSC, CD209, PPARγ, AKAP12, MAOA, WNT5A, STAT6, CCL18, and PCSK5 (panels A through L, respectively). Expression levels relative to resting macrophages (M0) in macrophages exposed to 20 ng/mL of IFN-γ plus 10 ng/mL of LPS in the presence (□) or absence (■) of the c-MYC inhibitor 10058-F4 (60μM) of CCR7 and CD80 (panels M and N, respectively). Results are shown as means ± SD of n = 5 independent experiments.
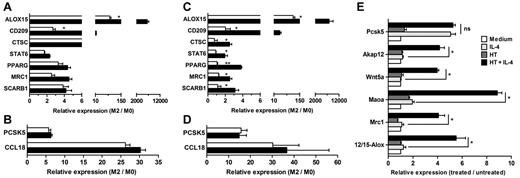
As a first approach to defining whether c-MYC induction was needed for the regulation of alternative activation genes, macrophages were treated with the protein synthesis inhibitor cycloheximide during IL-4 exposure, and the induction of c-MYC–dependent and c-MYC–independent genes was monitored for 24 hours by Q-PCR. Under these experimental conditions, induction by IL-4 of the c-MYC–dependent genes ALOX15 and CD209 was significantly affected, whereas other c-MYC–dependent genes were significantly more resistant to cycloheximide treatment, suggesting that basal levels of c-MYC may suffice for their expression (Figure 4A). Interestingly, the c-MYC–independent genes PSCK5 and CCL18 were unaffected (Figure 4B). The role of c-MYC was then further investigated by silencing c-MYC using a lentivirus-based shRNA (c-MYC/shRNA), which specifically and efficiently ablated c-MYC induction by IL-4 (supplemental Figure 1). Whereas macrophages infected with a mock lentivirus (mock/shRNA) responded to IL-4 similarly to uninfected macrophages, those infected with c-MYC/shRNA showed a significant inhibition of a set of alternative activation genes also inhibited by the c-MYC chemical inhibitor 10058-F4 (Figure 4C). The induction of genes not affected by c-MYC blockade, such as CCL18 and PCSK5, was also not significantly influenced by macrophage infection with c-MYC/shRNA (Figure 4D). Finally, further evidence for the role of c-Myc in the regulation of alternative activation genes was obtained using macrophages isolated from conditional gene–targeted c-Myc–fl/c-Myc–ERKI mice, in which c-Myc expression is controlled by the c-Myc endogenous promoter, but its activity, after deletion of the c-Myc floxed allele by exposure to Cre recombinase, is entirely dependent on the activation of the 4-HT–switchable form of the c-Myc protein (c-Myc-ERTAM) induced by exposure to the exogenous agonist 4-HT.32,33 Consistent with results obtained with the chemical inhibitor 10058-F4 and c-MYC/shRNA, in the absence of 4-HT (and hence c-Myc activity), IL-4–induced expression of Pcsk5 was preserved (Figure 4E). Conversely, the concomitant exposure to 4-HT was required to restore the inducibility of 12/15-Alox, Mrc1, Maoa, Wnt5a, and Akap12 by IL-4 (Figure 4E), demonstrating the requirement of c-Myc for the IL-4–dependent induction of these genes in alternatively activated macrophages.
Inhibition of c-MYC expression blocks the induction of c-MYC–dependent M2 markers. (A-B) Effect of cycloheximide (open bars) and vehicle (closed bars) on the induction of genes associated with alternative activation in human macrophages exposed to 20 ng/mL of IL-4 for 24 hours. Results are shown as means ± SD of n = 3 independent experiments. (C-D) Effect of c-MYC/shRNA (open bars) and mock/shRNA (closed bars) on the induction of genes associated with alternative activation in human macrophages exposed to 20 ng/mL of IL-4 for 24 hours. Results are shown as means ± SD of n = 3 independent experiments. (E) Expression levels of 12/15-Alox, Mcr1, Maoa, Wnt5a, Apak12, and Pcsk5 transcripts evaluated by Q-PCR in peritoneal murine macrophages isolated from c-Myc-fl/c-Myc-ERKI mice exposed to adeno-cre for 72 hours after stimulation for 24 hours with medium, 20 ng/mL of IL-4, 100nM 4-HT, or 20 ng/mL of IL-4 plus 100nM 4-HT. Results are shown as means ± SD of n = 3 independent experiments with 3 animals used in each experiment.
Inhibition of c-MYC expression blocks the induction of c-MYC–dependent M2 markers. (A-B) Effect of cycloheximide (open bars) and vehicle (closed bars) on the induction of genes associated with alternative activation in human macrophages exposed to 20 ng/mL of IL-4 for 24 hours. Results are shown as means ± SD of n = 3 independent experiments. (C-D) Effect of c-MYC/shRNA (open bars) and mock/shRNA (closed bars) on the induction of genes associated with alternative activation in human macrophages exposed to 20 ng/mL of IL-4 for 24 hours. Results are shown as means ± SD of n = 3 independent experiments. (E) Expression levels of 12/15-Alox, Mcr1, Maoa, Wnt5a, Apak12, and Pcsk5 transcripts evaluated by Q-PCR in peritoneal murine macrophages isolated from c-Myc-fl/c-Myc-ERKI mice exposed to adeno-cre for 72 hours after stimulation for 24 hours with medium, 20 ng/mL of IL-4, 100nM 4-HT, or 20 ng/mL of IL-4 plus 100nM 4-HT. Results are shown as means ± SD of n = 3 independent experiments with 3 animals used in each experiment.
Direct and indirect regulation of macrophage gene expression by c-MYC
ALOX15 and MRC1 have not been associated previously with c-MYC, whereas other alternative activation genes, including SCARB1 and CTSC, have been described previously as c-MYC target genes in different cell lines and primary cells.38 To further investigate the interaction of c-MYC with c-MYC–dependent alternative activation genes in macrophages, ChIP experiments were performed. Results confirmed a direct interaction of c-MYC with the SCARB1 promoter,38 and revealed its interaction with the ALOX15, PPARγ, and STAT6 promoters, which contained putative c-MYC–responsive elements (Figure 5A-D). Although it was sensitive to c-MYC inhibition (Figures 3E and 4C), CD209 did not contain putative c-MYC–responsive elements in its promoter and showed no direct c-MYC binding in ChIP experiments (Figure 5E). We hypothesize that CD209 dependency by c-MYC could be related to the induction by c-MYC of STAT6, which has putative binding sites within the CD209 promoter (Figure 5E). Finally, no putative c-MYC–responsive elements were predicted in the PCSK5 promoter region, which, consistent with its insensitivity to c-MYC inhibition (Figures 3L and 4D), did not show interaction with c-MYC in ChIP experiments (Figure 5F). To further investigate the regulation of c-MYC–dependent alternative activation genes, the role of the IL-4 transducers STAT6 and PPARγ was evaluated using a JAK2/3 inhibitor (AG490; 10μM) and a PPARγ antagonist (GW9662; 10μM), in comparison with the c-MYC inhibitor 10058-F4. Results indicate that regulation of the c-MYC–dependent genes SCARB1, ALOX15, and STAT6 (Figure 5G-I, respectively) does not involve these transcription factors, whereas STAT6 is involved in the induction by c-MYC of PPARγ and CD209 (Figure 5J-K, respectively). These results suggest that among c-MYC–dependent alternative activation genes, a distinct subset is controlled directly by c-MYC, whereas a second subset is influenced indirectly by c-MYC, possibly in part as a consequence of the impact of c-MYC on the expression of IL-4 signal transducers.
Role of c-MYC, STAT6, and PPARγ on the induction of c-MYC–sensitive M2 markers. Bioinformatic analysis identified candidate binding sites for c-MYC (CACGTG E-box sequence; ●) and STAT6 (TTCN3-4GAA sequence; ○) in the putative promoter region of SCARB1 (c-MYC sites: −198/−202 and −765/−770; A); ALOX15 (c-MYC sites: +295/+301 and −1318/−1323; B); STAT6 (c-MYC sites: −273/−279, −798/−803, −1198/−1203, −1873/−1878; C); PPARγ (c-MYC sites: −224/−229; D); and CD209 (STAT6 sites: −408/−417, −1521/−1540, −1585/−1594; (E). No c-MYC- or STAT6-binding sites were identified in PCSK5 (F). ChIP experiments performed with an anti–c-MYC Ab (closed bars) and an isotypic control (open bars) on the promoter region of indicated genes from unstimulated (M0) and IL-4–stimulated macrophages (M2) confirmed c-MYC binding in the putative promoter region of SCARB1, ALOX15, STAT6, and PPARγ (A- D). No evidence for c-MYC binding in the promoter region of CD209 (E) or PCSK5 (F) was detected. Results are shown as means ± SD of n = 3 independent experiments. To evaluate the role of c-MYC, STAT6, and PPARγ on the induction of the alternative activation genes, the c-MYC inhibitor 10058-F4 (60μM), the JAK2/3 inhibitor AG490 (10μM), and the PPARγ antagonist GW9662 (10μM) or their respective vehicles were added to macrophage cultures during polarization with IL-4 (M2). After 24 hours, expression levels of SCARB1, ALOX15, STAT6, PPARγ, CD209, and PCSK5 (panels G through L, respectively) were evaluated by Q-PCR. Results are shown as means ± SD of n = 3 independent experiments.
Role of c-MYC, STAT6, and PPARγ on the induction of c-MYC–sensitive M2 markers. Bioinformatic analysis identified candidate binding sites for c-MYC (CACGTG E-box sequence; ●) and STAT6 (TTCN3-4GAA sequence; ○) in the putative promoter region of SCARB1 (c-MYC sites: −198/−202 and −765/−770; A); ALOX15 (c-MYC sites: +295/+301 and −1318/−1323; B); STAT6 (c-MYC sites: −273/−279, −798/−803, −1198/−1203, −1873/−1878; C); PPARγ (c-MYC sites: −224/−229; D); and CD209 (STAT6 sites: −408/−417, −1521/−1540, −1585/−1594; (E). No c-MYC- or STAT6-binding sites were identified in PCSK5 (F). ChIP experiments performed with an anti–c-MYC Ab (closed bars) and an isotypic control (open bars) on the promoter region of indicated genes from unstimulated (M0) and IL-4–stimulated macrophages (M2) confirmed c-MYC binding in the putative promoter region of SCARB1, ALOX15, STAT6, and PPARγ (A- D). No evidence for c-MYC binding in the promoter region of CD209 (E) or PCSK5 (F) was detected. Results are shown as means ± SD of n = 3 independent experiments. To evaluate the role of c-MYC, STAT6, and PPARγ on the induction of the alternative activation genes, the c-MYC inhibitor 10058-F4 (60μM), the JAK2/3 inhibitor AG490 (10μM), and the PPARγ antagonist GW9662 (10μM) or their respective vehicles were added to macrophage cultures during polarization with IL-4 (M2). After 24 hours, expression levels of SCARB1, ALOX15, STAT6, PPARγ, CD209, and PCSK5 (panels G through L, respectively) were evaluated by Q-PCR. Results are shown as means ± SD of n = 3 independent experiments.
c-MYC is not involved in macrophage proliferation and survival
Because c-MYC is a potent effector of cell-cycle progression and apoptosis, we investigated its potential involvement in macrophage survival and proliferation. However, we did not obtain evidence for any effect of c-MYC inhibition on macrophage survival or on the cell cycle of human macrophages in our experimental conditions (supplemental Figure 2).
c-MYC is expressed in TAMs and controls expression of protumoral genes
In most tumor types, TAMs in several respects resemble several aspects of alternatively activated macrophages,8,9 although the existence of TAM subsets with distinct functional properties has been reported recently.8,9 In the tumor microenvironment, products derived either from the tumor30,39 or from the host, including IL-4,9,40,41 contribute to skewing macrophage function. It was therefore important to investigate the role of c-MYC in the biology of TAMs. TAMs can be mimicked in vitro by exposing macrophages to tumoral cell line–conditioned culture medium.30,39 In this simplified cell system, macrophage exposure to tumoral cell line–conditioned culture medium resulted in the induction of c-MYC (Figure 6A), as well as alterative activation markers including ALOX15 and MRC1 (Figure 6B-C). Tumor-conditioned medium also induced the expression of TGF-β, VEGF, the hypoxia inducible factor 1 α-subunit (HIF-1α), and matrix metalloproteinase 9 (MMP9). Furthermore, inhibition of c-MYC activity by treatment with its chemical inhibitor 10058-F4 or suppression of c-MYC expression by macrophage transduction with c-MYC/shRNA significantly inhibited the induction of these genes by tumor-conditioned medium (Figure 6B and C, respectively). Conversely, and in agreement with its identification as a c-MYC–independent alternative activation gene (Figure 3H), the induction of CCL18 by tumor-conditioned medium was unaffected by c-MYC blockade (induction 45.55 ± 8.56-fold vs 47.88 ± 9.04-fold in untreated and 10058-F4 treated macrophages, respectively, and 48.58 ± 15.31-fold vs 49.74 ± 10.98-fold in mock and c-MYC/shRNA-treated macrophages, respectively). Finally, immunohistochemical analysis detected c-MYC expression in a large fraction of CD68+ tumor-associated macrophages in human colon tumor specimens (Figure 6D), but c-MYC was not detected in CD68+ macrophages localized in the lamina propria of normal colon mucosa (Figure 6E), showing that c-MYC is expressed in vivo in human TAMs and not in resting macrophages.
c-Myc is expressed in human TAMs and regulates their phenotype. (A) c-Myc expression in human macrophages cultured or not in the presence of 20 ng/mL of IL-4 (M2 and M0, respectively) or culture medium conditioned by the human tumoral cell lines PANC-1 (pancreas tumor), SW480 (colon tumor), and MDAMB231 (breast tumor). Results are shown as means ± SD of n = 6 independent experiments. (B-C) Expression levels of ALOX15, MRC1, HIF1α, MMP9, TGFβ, and VEGF by single Q-PCR in human macrophages exposed for 24 hours to PANC-1–conditioned medium (B-C closed bars), in the presence of the c-MYC inhibitor 10058-F4 (60μM) (B open bars) or after suppression of c-MYC expression by macrophage transduction with a c-MYC/shRNA (C open bars). Results are the the -fold induction compared with untreated macrophages and represent the means ± SD of n = 4 independent experiments. (D-E) Double immunohistochemical analysis for CD68 (alkaline phosphatase staining: red color) and c-MYC (peroxidase staining: brown color) in specimens of colon cancer (D) and normal colon (E). Hematoxylin counterstaining. Magnification is 20×. In panel D, c-MYC expression is found in neoplastic cells (nuclear pattern, **) and in CD68+ macrophages of the lamina propria (double immunostaining: red and brown). A few c-MYC−/CD68+ macrophages are found (*). **Neoplastic glands; arrows, c-MYC+/CD68+ macrophages; *c-MYC−/CD68+ macrophages. In panel E, CD68+ macrophages, localized in the lamina propria of normal colon mucosa show no c-MYC immunostaining. **Normal colon glands; arrows, CD68+ macrophages. In both panels, inserts show high-magnification fields (100×) of c-MYC/CD68 immunostaining.
c-Myc is expressed in human TAMs and regulates their phenotype. (A) c-Myc expression in human macrophages cultured or not in the presence of 20 ng/mL of IL-4 (M2 and M0, respectively) or culture medium conditioned by the human tumoral cell lines PANC-1 (pancreas tumor), SW480 (colon tumor), and MDAMB231 (breast tumor). Results are shown as means ± SD of n = 6 independent experiments. (B-C) Expression levels of ALOX15, MRC1, HIF1α, MMP9, TGFβ, and VEGF by single Q-PCR in human macrophages exposed for 24 hours to PANC-1–conditioned medium (B-C closed bars), in the presence of the c-MYC inhibitor 10058-F4 (60μM) (B open bars) or after suppression of c-MYC expression by macrophage transduction with a c-MYC/shRNA (C open bars). Results are the the -fold induction compared with untreated macrophages and represent the means ± SD of n = 4 independent experiments. (D-E) Double immunohistochemical analysis for CD68 (alkaline phosphatase staining: red color) and c-MYC (peroxidase staining: brown color) in specimens of colon cancer (D) and normal colon (E). Hematoxylin counterstaining. Magnification is 20×. In panel D, c-MYC expression is found in neoplastic cells (nuclear pattern, **) and in CD68+ macrophages of the lamina propria (double immunostaining: red and brown). A few c-MYC−/CD68+ macrophages are found (*). **Neoplastic glands; arrows, c-MYC+/CD68+ macrophages; *c-MYC−/CD68+ macrophages. In panel E, CD68+ macrophages, localized in the lamina propria of normal colon mucosa show no c-MYC immunostaining. **Normal colon glands; arrows, CD68+ macrophages. In both panels, inserts show high-magnification fields (100×) of c-MYC/CD68 immunostaining.
Discussion
c-MYC is a pleiotropic transcription factor affecting somatic and germline cells in a variety of biologic processes, including cell proliferation, biosynthetic metabolism, and apoptosis.17,18 Because of its oncogenic potential, the biologic properties of c-MYC have been evaluated mostly in tumoral cells,42 and its functions on nontumoral immune cells have not been well characterized.43,44 In the immune compartment, c-MYC is known to play a role in the control of lymphocyte homeostasis and survival/apoptosis of myeloid cells.26,27 In the present study, we report that this transcription factor is induced in human macrophages during alternative activation and controls a significant fraction of genes associated with alternative activation, either through direct gene induction or via its weak effect on other known IL-4 key transcription factors.
ChIP experiments demonstrated a direct interaction of c-MYC with promoters of prototypic alternative activation genes, including ALOX15 and MRC1, and confirmed previous reports on direct c-MYC interaction with the SCARB1 promoter.38 Conversely, ALOX15 induction by IL-4 in various cell types, including primary cultures of human monocytes, had been suggested previously to be a STAT6-dependent event.45,46 Nonetheless, in T-cell lymphoma cells, histone deacetylase inhibitors able to reduce STAT6 and phospho-STAT6 protein expression did not affect ALOX15 expression,47 suggesting the existence of STAT6-independent intracellular pathways regulating ALOX15 expression in response to IL-4. Similarly, MRC1 induction by IL-4 in macrophages and dendritic cells has been well established.48 Interestingly, bioinformatic analysis did not reveal putative STAT6-binding sites within the MRC1 putative promoter, whereas multiple c-MYC–binding sites in the putative promoter regions of these M2 genes were identified and ChIP assays demonstrated their direct binding to c-MYC. Although the concomitant involvement of STAT6 cannot be excluded, our data highlight the direct involvement of c-MYC in the induction of a distinct subset of genes in macrophages exposed to IL-4, including the prototypic alternative activation genes ALOX15, MRC1, and SCARB1. In some c-MYC–dependent alternative activation genes, bioinformatic analysis did not predict c-MYC–binding sites, raising the possibility that, in some cases, c-MYC dependency could be related to the regulation of secondary transcription factors. Interestingly, among c-MYC–dependent alternative activation genes, we identified STAT6, which has been described previously as a c-MYC–dependent protein in other cell types.37 STAT6 and not c-MYC–binding sites were identified on the putative promoter of the c-MYC–dependent gene CD209, which did not show evidence of direct interaction with c-MYC by ChIP analysis. These results are consistent with an indirect involvement of c-MYC in the regulation of M2 genes that do not have c-MYC–binding sites within their promoter, including CD209, via a STAT6-dependent amplification loop. Although we have no evidence from the panel of alternative activation genes investigated, a similar scenario could apply to PPARγ, a transcription factor known to be required for M2 phenotype acquisition in murine macrophages.49 PPARγ also has c-MYC–binding sites in its putative promoter region, and in ChIP analysis showed direct interaction with c-MYC. In alternative macrophages, c-MYC controls both PPARγ and ALOX15, which have been involved in the generation and activity of endogenous ligands controlling a set of alternative activation genes.50 Finally, c-MYC–binding sites have not been predicted in the putative promoter regions of c-MYC–insensitive M2 genes, including CCL18 and PCSK5, and ChIP analysis failed to show direct interaction of c-MYC with these M2 genes. Therefore, the transcriptome of alternatively activated macrophages includes approximately 55% of genes regulated independently of c-MYC. Among the 45% M2 genes showing significant and reproducible dependency on c-MYC, we have identified genes regulated by direct interaction of c-MYC (ALOX15, MRC1, and SCARB1) and genes controlled by c-MYC through indirect pathways likely involving the regulation by c-MYC of other master transcription factors operating in alternatively activated macrophages, such as STAT6 and PPARγ.
In tumors, macrophages are exposed to various signals derived from the tumor microenvironment, such as soluble molecules and cell-cell interactions, which contribute to the acquisition of the TAM phenotype, which has several similarities with the M2 phenotype.9,30 Interestingly, c-MYC was induced in macrophages exposed in vitro to conditioned medium from different human tumor cell lines and was detected in CD68+ macrophages infiltrating human colon tumors. These results suggest a general mechanism of c-MYC up-regulation in human TAMs in different tumor microenvironments. As in alternatively activated macrophages, in human tumor-conditioned macrophages, c-MYC activity was required for ALOX15 and MRC1 expression and was also involved in the expression of genes with a relevant role in TAM biology, including HIF-1α, TGF-β, MMP9, and VEGF.51 These results reveal an important role of c-MYC in the control of protumoral properties of TAMs, and suggest that the ability of c-MYC inhibitors to interfere with TAM activation should be taken into account when assessing their antitumoral activity.34
We conclude that c-MYC is a key element in alternative macrophage activation, being involved in the induction of a significant set of alternative activation genes, either directly (eg, MRC1 and ALOX15) or indirectly, likely through its positive effect on other key transcription factors (eg, STAT6 and PPARγ). c-MYC is involved in TAM regulation, where it controls the expression of key mediators (MMP9, VEGF, HIF-1α, and TGF-β). The results of the present study identify c-MYC as a key regulator in macrophage biology and support the use of this transcription factor for therapeutic approaches targeting pathologies associated with alternatively activated macrophages, including tumors.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by research grants from the European Community (INNOCHEM project 518167), the Ministero dell'Istruzione dell'Università e della Ricerca (PRIN projects 2002061255 and 2007-ENYMAN_003; FIRB project RBFR08CW8G), the Alleanza Contro il Cancro and Istituto Superiore di Sanità (Programma Straordinario di Ricerca Oncologica 2006-RNBIO project), the Italian Association for Cancer Research (AIRC), Regione Lombardia (LIIN project), and Fondazione Cariplo (NOBEL project). This work was conducted in the context of Fondazione Humanitas per la Ricerca. O.M.P. is the recipient of a fellowship from Fundación Alfonso Martín Escudero. L.Z. is the recipient of a fellowship from Fondazione Cariplo.
Authorship
Contribution: O.M.P. designed the research, performed the experiments, analyzed the results, produced the figures, and wrote the manuscript; M.D.P., M.M., A.A., and A.D. performed the in vitro experiments and analyzed the results; L.Z. performed the microarray experiments and analyzed the results; L.S., L.B.S., and G.I.E. performed the in vivo experiments and analyzed the results; M.N. performed the immunohistochemical analysis and analyzed the results; and A.M. and M.L. designed the research, analyzed the results, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for O.M.P. is Department of Epidemiology, Atherothrombosis and Cardiovascular Imaging, Centro Nacional de Investigaciones Cardiovasculares, Madrid, Spain.
Correspondence: Massimo Locati, MD, PhD, Department of Translational Medicine, University of Milan, Istituto Clinico Humanitas IRCCS, Via Manzoni 56, I-20089 Rozzano, Italy; e-mail: massimo.locati@humanitasresearch.it.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal