Abstract
A cohort of MDS patients was examined for mutations affecting 4 splice genes (SF3B1, SRSF2, ZRSR2, and U2AF35) and evaluated in the context of clinical and molecular markers. Splice gene mutations were detected in 95 of 221 patients. These mutations were mutually exclusive and less likely to occur in patients with complex cytogenetics or TP53 mutations. SF3B1mut patients presented with lower hemoglobin levels, increased WBC and platelet counts, and were more likely to have DNMT3A mutations. SRSF2mut patients clustered in RAEB-1 and RAEB-2 subtypes and exhibited pronounced thrombocytopenias. ZRSR2mut patients clustered in International Prognostic Scoring System intermediate-1 and intermediate-2 risk groups, had higher percentages of bone marrow blasts, and more often displayed isolated neutropenias. SRSF2 and ZRSR2 mutations were more common in TET2mut patients. U2AF35mut patients had an increased prevalence of chromosome 20 deletions and ASXL1 mutations. Multivariate analysis revealed an inferior overall survival and a higher AML transformation rate for the genotype ZRSR2mut/TET2wt (overall survival: hazard ratio = 3.3; 95% CI, 1.4-7.7; P = .006; AML transformation: hazard ratio = 3.6; 95% CI, 2-4.2; P = .026). Our results demonstrate that splice gene mutations are among the most frequent molecular aberrations in myelodysplastic syndrome, define distinct clinical phenotypes, and show preferential associations with mutations targeting transcriptional regulation.
Introduction
Myelodysplastic syndromes (MDSs) are a heterogeneous group of myeloid neoplasms showing clonal hematopoiesis, aberrant differentiation, peripheral cytopenias, and risk of progression to acute myeloid leukemia (AML).1 Although cytopenias are the major clinical challenge in low-risk disease, transformation to AML can be observed in a significant number of high-risk MDS patients. The broad range of individual genes affected by mutations indicates that a variety of molecular mechanisms are involved in the pathogenesis of these disorders. Like TET2,2,3 the most commonly affected gene in MDS, several others, such as ASXL1,4 EZH2,5,6 and DNMT3A,7 are involved in epigenetic regulation of transcription. Additional genes known to be mutated in MDS include RUNX1,8 IDH1/2,9 TEL/ETV6,8 TP53,8 and NRAS.8 As MDS patient outcome remains dismal, identification of novel molecular markers in MDS that allows further subclassification and possibly risk-directed therapeutic intervention remains of major interest.8,10 Next-generation sequencing approaches have identified mutations in genes encoding multiple components of the RNA splicing machinery. The most frequently documented mutations affect splice genes U2AF35, ZRSR2, SRSF2, and SF3B1. Collectively, these mutations were observed in up to 85% of different myeloid neoplasms with myelodysplastic features,11,12 thus suggesting an important contribution of genetic alterations involving splicing components to the pathophysiology of MDS.
The detailed splicing mechanism is complex and relies on 5 small nuclear ribonucleoproteins (snRNP) and their associated proteins, which form the spliceosome. It processes premessenger RNA (pre-mRNA) into mature mRNA and controls the diversity of splice variants.13 Each snRNP is composed of a single uridine-rich small nuclear RNA (snRNA) accompanied by multiple proteins and has a specific function. The recently discovered mutations in the splicing machinery of patients with myeloid neoplasms are predicted to affect core components of initial steps, such as the recognition of the 3′ splice acceptor site of the pre-mRNA target intron (U2AF35 and SRSF2) or the recruitment of the U2 snRNP to the branch point proximal to the 3′ splicing site that contains SF3B1. Although abnormal splicing was described to be associated with U2AF3511,14 and SF3B112 mutations, the biologic mechanisms linking the splicing machinery to cellular transformation and leukemogenesis remain elusive. Since the initial report on SF3B1 mutations in MDS, other groups have confirmed the high frequency of SF3B1 mutations in MDS patients and the association of the mutations with the presence of ring sideroblasts (RSs).15-17 However, little is currently known about the clinical course, morphologic features, prognostic impact, and concomitant molecular aberrations of MDS patients harboring SRSF2, ZRSR2, or U2AF35 mutations.
Therefore, we examined genomic DNA of 221 MDS patients at diagnosis for the presence of SF3B1, SRSF2, ZRSR2, and U2AF35 mutations by direct sequencing and evaluated their prognostic impact with regard to other mutations, including ASXL1, CBL, DNMT3A, ETV6, EZH2, IDH1/2, JAK2, NRAS, RUNX1, TET2, and TP53.
Methods
Patients
The 221 MDS samples were collected at time of enrolment in multicenter clinical trials in France between 1999 and 2011 at Paris Cochin (n = 134) and Marseille Institute Paoli-Calmette (n = 87). Clinical and hematologic data were recorded after informed consent in accordance with the Declaration of Helsinki, and the analysis of the samples was approved by the institutional review boards of Paris Centre and Marseille. The distribution of WHO subtypes, International Prognostic Scoring System (IPSS) risk groups, and cytogenetic risk groups (according to WHO 200818 ) is shown in Table 1. Follow-up information was available for 198 of the 221 MDS patients. The follow-up information was updated by means of clinic visits. Treatment details for both cohorts have been published earlier4,19 and are shown according to mutation status in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Clinical characteristics of 221 MDS patients according to SF3B1, SRSF2, ZRSR2, and U2AF35 mutation status
| Characteristic . | All . | SF3B1 mut (n = 37) . | SF3B1 wt (n = 184) . | P . | SRSF2 mut (n = 25) . | SRSF2 wt (n = 196) . | P . | ZRSR2 mut (n = 25) . | ZRSR2 wt (n = 196) . | P . | U2AF35 mut (n = 12) . | U2AF35 wt (n = 209) . | P . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age, y | 217 | .17 | .25 | .7 | .71 | ||||||||
| Median | 71.9 | 73 | 71 | 70 | 72 | 71.9 | 71.9 | 73.5 | 71.9 | ||||
| Range | 51-95 | 35-95 | 49-87.8 | 35-95 | 57.9-95 | 35-95 | 56-88 | 35-95 | |||||
| Sex | 221 | .88 | 0.86 | < .001 | .016 | ||||||||
| Male, no. (%) | 129 | 22 (59) | 107 (58) | 15 (60) | 114 (58) | 24 (96) | 105 (54) | 11 (92) | 118 (56) | ||||
| WHO subtype | 221 | < .001 | .19 | .048 | .48 | .48 | |||||||
| RA, no. (%) | 34 | 2 (5) | 32 (17) | 2 (8) | 32 (17) | 1 (4) | 33 (17) | 4 (33) | 30 (14) | ||||
| RARS, no. (%) | 27 | 20 (54) | 7 (4) | 1 (4) | 26 (13) | 0 (0) | 27 (14) | 1 (8) | 26 (13) | ||||
| RCMD, no. (%) | 31 | 0 (0) | 31 (17) | 3 (12) | 28 (14) | 3 (12) | 28 (14) | 1 (8) | 30 (14) | ||||
| RCMD-RS, no. (%) | 8 | 6 (16) | 2 (1) | 0 (0) | 8 (4) | 0 (0) | 8 (4) | 1 (8) | 7 (3) | ||||
| RAEB-1, no. (%) | 55 | 1 (3) | 54 (29) | 12 (48) | 43 (22) | 11 (44) | 44 (23) | 1 (8) | 54 (26) | ||||
| RAEB-2, no. (%) | 45 | 3 (8) | 42 (23) | 6 (24) | 39 (20) | 9 (36) | 36 (18) | 4 (33) | 41 (20) | ||||
| 5q− syndrome, no. (%) | 2 | 0 (0) | 2 (1) | 0 (0) | 2 (1) | 0 (0) | 2 (1) | 0 (0) | 2 (1) | ||||
| RARS-T, no. (%) | 6 | 5 (14) | 1 (1) | 0 (0) | 6 (3) | 0 (0) | 6 (3) | 0 (0) | 6 (3) | ||||
| MDS-U, no. (%) | 13 | 0 (0) | 13 (7) | 1 (4) | 12 (6) | 1 (4) | 12 (6) | 0 (0) | 13 (6) | ||||
| RSs | < .001 | .047 | .011 | .86 | |||||||||
| Present, no. (%) | 41 | 31 (84) | 10 (5) | 1 (4) | 40 (20) | 0 (0) | 41 (21) | 2 (17) | 39 (19) | ||||
| Karyotype risk | 213 | .71 | .21 | .25 | .99 | ||||||||
| Low, no. (%) | 155 | 28 (76) | 128 (70) | 15 (60) | 141 (72) | 20 (80) | 136 (69) | 9 (75) | 147 (70) | ||||
| Intermediate, no. (%) | 38 | 7 (19) | 31 (17) | 7 (28) | 31 (16) | 5 (20) | 33 (17) | 2 (17) | 36 (17) | ||||
| High, no. (%) | 19 | 2 (5) | 17 (9) | 1 (4) | 18 (9) | 0 (0) | 19 (10) | 1 (8) | 18 (9) | ||||
| Bone marrow blasts | 217 | < .001 | .022 | .011 | .77 | ||||||||
| Median, % | 4 | 2 | 5 | 5 | 4 | 6.5 | 4 | 3 | 4 | ||||
| Range, % | 1-19 | 0-19 | 1-18 | 0-19 | 1-18 | 0-19 | 1-16 | 0-19 | |||||
| Hemoglobin | 211 | < .001 | .13 | .7 | .54 | ||||||||
| Median, g/dL | 9.8 | 9 | 10.1 | 10.9 | 9.8 | 9.4 | 9.9 | 10.1 | 9.8 | ||||
| Range, g/dL | 6-11.8 | 6-15 | 7.1-15 | 6-15 | 7.7-15 | 6-15 | 7.5-14.5 | 6-15 | |||||
| WBC count | 207 | .001 | .046 | .008 | .44 | ||||||||
| Median, × 109/L | 4.2 | 6.1 | 4 | 3.2 | 4.3 | 2.9 | 4.3 | 5.2 | 4.2 | ||||
| Range, × 109/L | 1.8-36.7 | 0.9-18.4 | 1.2-13 | 0.9-36.7 | 0.9-6.8 | 1.2-36.7 | 1.8-8.5 | 0.9-36.7 | |||||
| Neutrophil count | 205 | .001 | .006 | .009 | .31 | ||||||||
| Median, × 109/L | 2.1 | 3.4 | 1.9 | 1.1 | 2.3 | 1.1 | 2.2 | 2.8 | 2.1 | ||||
| Range, × 109/L | 0.7-23.1 | 0.3-15.8 | 0.3-9.7 | 0.3-23.1 | 0.3-4.9 | 0.3-23.1 | 0.5-6.5 | 0.3-23.1 | |||||
| Platelet count | 212 | < .001 | < .001 | .4 | .47 | ||||||||
| Median, × 109/L | 156 | 271 | 141 | 88 | 165.5 | 149 | 157.5 | 134.5 | 156 | ||||
| Range, × 109/L | 23-1398 | 5-714 | 6-359 | 5-1398 | 48-290 | 5-1398 | 32-500 | 5-1398 | |||||
| IPSS, no. | 216 | .036 | .042 | .017 | .07 | ||||||||
| Low risk, % | 74 | 19 (51) | 55 (30) | 3 (12) | 71 (36) | 3 (12) | 71 (36) | 5 (42) | 69 (33) | ||||
| Intermediate-1, % | 91 | 15 (41) | 76 (41) | 11 (44) | 80 (41) | 16 (64) | 74 (38) | 2 (17) | 89 (43) | ||||
| Intermediate-2, % | 26 | 1 (3) | 25 (13) | 6 (24) | 20 (10) | 5 (20) | 21 (11) | 4 (33) | 22 (11) | ||||
| High, % | 25 | 2 (5) | 23 (13) | 4 (16) | 21 (11) | 1 (4) | 24 (12) | 1 (8) | 24 (11) | ||||
| Cytology | |||||||||||||
| Multilineage dysplasia, no. (%) | 115/204 | 14 (41) | 101 (59) | .050 | 17 (71) | 98 (54) | .13 | 21 (84) | 94 (53) | .003 | 5 (45) | 110 (57) | .45 |
| Dyserythropoiesis, no. (%) | 137/197 | 30 (94) | 107 (65) | .001 | 16 (70) | 121 (70) | 1 | 17 (68) | 120 (69) | .63 | 8 (73) | 129 (69) | .81 |
| Dysgranulopoiesis, no. (%) | 140/201 | 15 (47) | 125 (74) | .002 | 23 (92) | 117 (66) | .009 | 22 (88) | 118 (67) | .033 | 7 (64) | 133 (70) | .66 |
| Dysmegakayopoiesis, no. (%) | 131/200 | 14 (44) | 114 (68) | .008 | 21 (84) | 110 (63) | .038 | 20 (80) | 108 (62) | .038 | 7 (64) | 121 (64) | .96 |
| Transfusion dependence | 193 | .024 | .036 | .38 | .27 | ||||||||
| Yes, no. (%) | 91 | 22 (59) | 69 (38) | 5 (20) | 86 (44) | 8 (32) | 83 (42) | 3 (25) | 88 (42) |
| Characteristic . | All . | SF3B1 mut (n = 37) . | SF3B1 wt (n = 184) . | P . | SRSF2 mut (n = 25) . | SRSF2 wt (n = 196) . | P . | ZRSR2 mut (n = 25) . | ZRSR2 wt (n = 196) . | P . | U2AF35 mut (n = 12) . | U2AF35 wt (n = 209) . | P . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age, y | 217 | .17 | .25 | .7 | .71 | ||||||||
| Median | 71.9 | 73 | 71 | 70 | 72 | 71.9 | 71.9 | 73.5 | 71.9 | ||||
| Range | 51-95 | 35-95 | 49-87.8 | 35-95 | 57.9-95 | 35-95 | 56-88 | 35-95 | |||||
| Sex | 221 | .88 | 0.86 | < .001 | .016 | ||||||||
| Male, no. (%) | 129 | 22 (59) | 107 (58) | 15 (60) | 114 (58) | 24 (96) | 105 (54) | 11 (92) | 118 (56) | ||||
| WHO subtype | 221 | < .001 | .19 | .048 | .48 | .48 | |||||||
| RA, no. (%) | 34 | 2 (5) | 32 (17) | 2 (8) | 32 (17) | 1 (4) | 33 (17) | 4 (33) | 30 (14) | ||||
| RARS, no. (%) | 27 | 20 (54) | 7 (4) | 1 (4) | 26 (13) | 0 (0) | 27 (14) | 1 (8) | 26 (13) | ||||
| RCMD, no. (%) | 31 | 0 (0) | 31 (17) | 3 (12) | 28 (14) | 3 (12) | 28 (14) | 1 (8) | 30 (14) | ||||
| RCMD-RS, no. (%) | 8 | 6 (16) | 2 (1) | 0 (0) | 8 (4) | 0 (0) | 8 (4) | 1 (8) | 7 (3) | ||||
| RAEB-1, no. (%) | 55 | 1 (3) | 54 (29) | 12 (48) | 43 (22) | 11 (44) | 44 (23) | 1 (8) | 54 (26) | ||||
| RAEB-2, no. (%) | 45 | 3 (8) | 42 (23) | 6 (24) | 39 (20) | 9 (36) | 36 (18) | 4 (33) | 41 (20) | ||||
| 5q− syndrome, no. (%) | 2 | 0 (0) | 2 (1) | 0 (0) | 2 (1) | 0 (0) | 2 (1) | 0 (0) | 2 (1) | ||||
| RARS-T, no. (%) | 6 | 5 (14) | 1 (1) | 0 (0) | 6 (3) | 0 (0) | 6 (3) | 0 (0) | 6 (3) | ||||
| MDS-U, no. (%) | 13 | 0 (0) | 13 (7) | 1 (4) | 12 (6) | 1 (4) | 12 (6) | 0 (0) | 13 (6) | ||||
| RSs | < .001 | .047 | .011 | .86 | |||||||||
| Present, no. (%) | 41 | 31 (84) | 10 (5) | 1 (4) | 40 (20) | 0 (0) | 41 (21) | 2 (17) | 39 (19) | ||||
| Karyotype risk | 213 | .71 | .21 | .25 | .99 | ||||||||
| Low, no. (%) | 155 | 28 (76) | 128 (70) | 15 (60) | 141 (72) | 20 (80) | 136 (69) | 9 (75) | 147 (70) | ||||
| Intermediate, no. (%) | 38 | 7 (19) | 31 (17) | 7 (28) | 31 (16) | 5 (20) | 33 (17) | 2 (17) | 36 (17) | ||||
| High, no. (%) | 19 | 2 (5) | 17 (9) | 1 (4) | 18 (9) | 0 (0) | 19 (10) | 1 (8) | 18 (9) | ||||
| Bone marrow blasts | 217 | < .001 | .022 | .011 | .77 | ||||||||
| Median, % | 4 | 2 | 5 | 5 | 4 | 6.5 | 4 | 3 | 4 | ||||
| Range, % | 1-19 | 0-19 | 1-18 | 0-19 | 1-18 | 0-19 | 1-16 | 0-19 | |||||
| Hemoglobin | 211 | < .001 | .13 | .7 | .54 | ||||||||
| Median, g/dL | 9.8 | 9 | 10.1 | 10.9 | 9.8 | 9.4 | 9.9 | 10.1 | 9.8 | ||||
| Range, g/dL | 6-11.8 | 6-15 | 7.1-15 | 6-15 | 7.7-15 | 6-15 | 7.5-14.5 | 6-15 | |||||
| WBC count | 207 | .001 | .046 | .008 | .44 | ||||||||
| Median, × 109/L | 4.2 | 6.1 | 4 | 3.2 | 4.3 | 2.9 | 4.3 | 5.2 | 4.2 | ||||
| Range, × 109/L | 1.8-36.7 | 0.9-18.4 | 1.2-13 | 0.9-36.7 | 0.9-6.8 | 1.2-36.7 | 1.8-8.5 | 0.9-36.7 | |||||
| Neutrophil count | 205 | .001 | .006 | .009 | .31 | ||||||||
| Median, × 109/L | 2.1 | 3.4 | 1.9 | 1.1 | 2.3 | 1.1 | 2.2 | 2.8 | 2.1 | ||||
| Range, × 109/L | 0.7-23.1 | 0.3-15.8 | 0.3-9.7 | 0.3-23.1 | 0.3-4.9 | 0.3-23.1 | 0.5-6.5 | 0.3-23.1 | |||||
| Platelet count | 212 | < .001 | < .001 | .4 | .47 | ||||||||
| Median, × 109/L | 156 | 271 | 141 | 88 | 165.5 | 149 | 157.5 | 134.5 | 156 | ||||
| Range, × 109/L | 23-1398 | 5-714 | 6-359 | 5-1398 | 48-290 | 5-1398 | 32-500 | 5-1398 | |||||
| IPSS, no. | 216 | .036 | .042 | .017 | .07 | ||||||||
| Low risk, % | 74 | 19 (51) | 55 (30) | 3 (12) | 71 (36) | 3 (12) | 71 (36) | 5 (42) | 69 (33) | ||||
| Intermediate-1, % | 91 | 15 (41) | 76 (41) | 11 (44) | 80 (41) | 16 (64) | 74 (38) | 2 (17) | 89 (43) | ||||
| Intermediate-2, % | 26 | 1 (3) | 25 (13) | 6 (24) | 20 (10) | 5 (20) | 21 (11) | 4 (33) | 22 (11) | ||||
| High, % | 25 | 2 (5) | 23 (13) | 4 (16) | 21 (11) | 1 (4) | 24 (12) | 1 (8) | 24 (11) | ||||
| Cytology | |||||||||||||
| Multilineage dysplasia, no. (%) | 115/204 | 14 (41) | 101 (59) | .050 | 17 (71) | 98 (54) | .13 | 21 (84) | 94 (53) | .003 | 5 (45) | 110 (57) | .45 |
| Dyserythropoiesis, no. (%) | 137/197 | 30 (94) | 107 (65) | .001 | 16 (70) | 121 (70) | 1 | 17 (68) | 120 (69) | .63 | 8 (73) | 129 (69) | .81 |
| Dysgranulopoiesis, no. (%) | 140/201 | 15 (47) | 125 (74) | .002 | 23 (92) | 117 (66) | .009 | 22 (88) | 118 (67) | .033 | 7 (64) | 133 (70) | .66 |
| Dysmegakayopoiesis, no. (%) | 131/200 | 14 (44) | 114 (68) | .008 | 21 (84) | 110 (63) | .038 | 20 (80) | 108 (62) | .038 | 7 (64) | 121 (64) | .96 |
| Transfusion dependence | 193 | .024 | .036 | .38 | .27 | ||||||||
| Yes, no. (%) | 91 | 22 (59) | 69 (38) | 5 (20) | 86 (44) | 8 (32) | 83 (42) | 3 (25) | 88 (42) |
Cytogenetic and mutation analysis
Cytogenetic analysis was performed by G- and R-banding analysis. Mononuclear cells from diagnostic bone marrow samples were enriched by Ficoll density gradient centrifugation and were stored in liquid nitrogen until use. Genomic DNA was extracted from samples using the All Prep DNA/RNA Kit (QIAGEN) according to the manufacturer's recommendations. Genomic DNA was amplified by linear whole genome amplification using the REPLI-G Kit (QIAGEN) to perform a first identification screen. All candidate mutations were subsequently analyzed in an independent experiment using nonamplified genomic DNA. The genomic regions that span exon 12 of ASXL1,4 exons 8 and 9 of CBL,20 exons 15 to 23 of DNMT3A,21 the entire coding regions of ETV6 and EZH2,6 exons 4 of IDH1 and IDH2,22 exon 14 of JAK2,23 exons 1 and 2 of NRAS,24 exons 3 to 8 of RUNX1,25 the entire coding region of TET2,19 and exons 5 to 8 of TP5325 were analyzed as previously reported. SF3B1 (exons 12-16), SRSF2 (exon2), ZRSR2 (exons 1-11), and U2AF35 (exons 2 and 6) were amplified using intron-flanking primers tagged with M13 universal primers at the 3′ or 5′ ends. PCR fragments were directly sequenced in both directions and were analyzed using the Mutation surveyor Version 3.97 software (Softgenetics). Nontumoral tissue was analyzed when available (DNA from buccal swab or CD3+ T cells; n = 20).
Statistical analysis
Overall survival (OS) end points, measured from the date of diagnosis, were death (failure) and alive at last follow-up (censored). Time to AML progression was measured from the date of MDS diagnosis to the time of AML diagnosis. Progression to AML was defined according to the 2008 WHO classification. The median follow-up time for patients alive was calculated according to the method of Korn.26 Primary analysis was performed on OS and time to AML progression. The Kaplan-Meier method, log-rank test, and Cox proportional hazards models were used to estimate the distribution of OS and time to AML progression and to compare differences between survival curves, respectively. Pairwise comparisons were performed by Mann-Whitney test for continuous variables and by 2-sided Fisher exact or χ2 tests for categorical variables, and are provided for exploratory purposes.
For multivariate analysis, a Cox proportional hazards model was constructed for OS and time to progression to AML, adjusting for potential confounding covariates.27 Variables considered for model inclusion were IPSS risk group, transfusion dependence, age (below vs above median), and mutation status of all 16 analyzed genes (mutated vs wild-type). Variables with P ≤ .1 in univariate analysis for OS were included in the model. The statistical analyses were performed with the statistical software package SPSS Version 19.0 (SPSS). Genomic alterations were considered as a mutation when the variation was not listed in dbSNP database (build 131 and 132), shown to be acquired or had been identified as such in other studies.
Results
Mutation status of SF3B1, SRSF2, ZRSR2, and U2AF35 in MDS patients
Among the 221 patients with MDS, 37 had SF3B1 mutations (16.4%), 25 had ZRSR2 mutations (11.1%), 25 had SRSF2 mutations (11.1%), and 12 patients harbored mutations affecting U2AF35 (5.4%). In total, 99 mutations affecting one of the 4 genes were detected in 95 patients (42.2%). Splice gene mutations were mostly mutually exclusive (91 of 95, 97%; P < .001), with concomitant mutations affecting 2 splice genes detected in only 4 patients. Two patients harbored mutations in both SRSF2 and ZRSR2, one patient had SF3B1 and SRSF2 mutations, and another patient had SF3B1 and ZRSR2 mutations.
The 37 mutations in SF3B1 were all heterozygous missense mutations affecting 6 mutational hotspots that are located in the HEAT domains 3 to 6 (Figure 1). The most common recurrent SF3B1 mutation affected amino acid residue K700 (21 of 37, 56.7%). The somatic nature of SF3B1 mutations was confirmed by sequencing nontumoral CD3+ cells in 8 patients (6 patients, K700E; 1 patient, D781G; and 1 patient, R625L). Except for detection of Y44H in one patient, all mutations observed in SRSF2 affected amino acid residue P95 and were predominantly heterozygous missense mutations. Notably, a 24-bp deletion starting at amino acid residue P95 was found in 3 patients. The somatic nature of this deletion was verified by sequencing CD3+ cells from one patient. Except for one frameshift mutation starting at Q157, mutations in U2AF35 affected positions S34 and Q157 and were heterozygous missense mutations. The acquired nature of substitutions S34F and Q157R was verified by sequencing DNA from CD3+ cells (n = 2) or buccal swab (n = 1). In contrast to SF3B1, SRSF2, and U2AF35, mutations in ZRSR2 were spread over the entire coding region and were mainly frameshift, splice site, or nonsense mutations (17 of 25, 68%). A 6-bp insertion starting at amino acid residue R446, resulting in the duplication of the amino acids arginine and serine, was observed in 9 patients (9 of 221, 4.1%). This duplication was also detected in nontumoral CD3+ cells of 2 patients and therefore classified as a polymorphism. A detailed overview of mutation sites is shown in Figure 1.
Localization of mutations identified in splice genes. Each mutation is shown with a circle. ● represents confirmed somatic mutations; and colored boxes, known domain structures. ZN indicates zinc finger; RRM, RNA recognition motif; RS, arginine-serine-rich domain; and HD, HEAT domain.
Localization of mutations identified in splice genes. Each mutation is shown with a circle. ● represents confirmed somatic mutations; and colored boxes, known domain structures. ZN indicates zinc finger; RRM, RNA recognition motif; RS, arginine-serine-rich domain; and HD, HEAT domain.
Clinical phenotype of patients harboring splice gene mutations
Patients with SF3B1 mutations presented with a distinct blood count at diagnosis. There was no significant difference in age, sex, or karyotype between SF3B1mut and SF3B1wt patients. Hemoglobin levels were significantly lower (median: 9 vs 10.1 g/dL; P < .001), while white blood cell counts (WBC; P = .001) and platelets (P < .001) were higher in SF3B1mut compared with SF3B1wt patients. Cytologic evaluation revealed a higher proportion of dysmorphic features in the erythroid lineage (94% vs 65%, P = .001), whereas multilineage dysplasia, dysgranulopoiesis, and dysmegakaryopoiesis were less often observed (Table 1). The percentage of bone marrow blasts was also lower in SF3B1mut patients (median: 2% vs 5%; P < .001). A strong association between the presence of RSs and SF3B1 mutation was observed: 31 of 41 patients with RS harbored a mutation in SF3B1 (84% vs 5%; P < .001). A bivariate regression analysis, including RS and SF3B1 mutation status, revealed an independent association for SF3B1mut patients with lower hemoglobin levels (odds ratio = 0.21, 95% CI, 0.06-0.7, P = .01). These findings resulted in a higher proportion of transfusion dependence for SF3B1mut compared with SF3B1wt patients (59% vs 38%; P = .024).
In contrast, SRSF2mut patients presented with substantially decreased numbers of neutrophil granulocytes (median: 1.1 vs 2.3 × 109/L; P = .006) and platelets (median: 88 vs 165 × 109/L; P < .001), whereas no differences for hemoglobin levels were observed. Dysplastic features affected predominantly granulopoiesis and megakaryopoiesis. Eighteen of 25 SRSF2mut patients were classified according to the WHO classification as refractory anemia with excess of blasts (RAEB-1) or RAEB-2 (72% vs 42% in SRSF2wt patients; P = .004). RSs were observed in one single SRSF2mut patient, and this case was the only one with a concomitant SF3B1 mutation. The transfusion dependence rate was significantly lower for SRSF2mut patients than for SRSF2wt patients (20% vs 44% in SRSF2wt patients; P = .036).
Patients harboring ZRSR2 mutations were almost exclusively male (24 of 25, 96%) and often presented with isolated neutropenias (median: 1.1 vs 2.2 × 109/L; P = .009). RSs were not observed in any of the 25 ZRSR2mut patients (P = .011). Twenty of 25 ZRSR2mut patients were classified according to the WHO classification as RAEB-1 or RAEB-2 (80% vs 41% in ZRSR2wt patients; P < .001, Table 1; supplemental Table 2). ZRSR2mut patients mainly clustered into the IPSS intermediate-1 and intermediate-2 risk groups (84% vs 49% in ZRSR2wt patients; P = .017).
Almost all U2AF35mut patients were male (11 of 12 = 92%; P = .016), but no significant differences in age, WHO classification, bone marrow blasts, hemoglobin, WBC, platelets, transfusion dependence, or IPSS score were observed. Interestingly, patients with a del20q were more likely to have U2AF35 mutations than patients with no del20q (30% vs 4.5%; P = .014).
Molecular associations of splice gene mutations in MDS
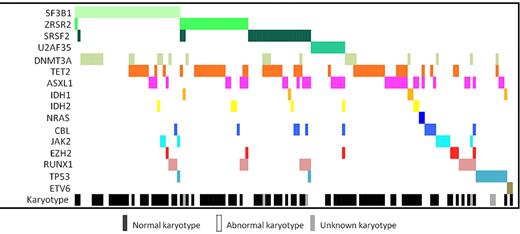
In our MDS patient cohort, TET2 mutations were the most common (54 of 221, 24.4%), whereas mutations in ASXL1 (37 of 221, 16.4%), DNMT3A (26 of 221, 11.6%), RUNX1 (16 of 221, 7.1%), TP53 (13 of 221, 5.8%), IDH1/2 (12 of 221, 5.3%), CBL (11 of 221, 4.9%), JAK2 (9 of 221, 4%), EZH2 (7 of 221, 3.1%), ETV6 (2 of 221, 0.9%), and NRAS (2 of 221, 0.9%) were less frequent. At least one mutation in any of the 16 analyzed genes was found in 154 of 221 patients (69.7%; Figure 2). Mutations in splice genes (n = 95 patients) were mutually exclusive from the presence of a complex karyotype (1.1% vs 12.6% in patients wild-type for the 4 splice genes; P = .001) and were significantly less often detected in patients with TP53 mutations (2% vs 9% in patients wild-type for the 4 splice genes; P = .04). To identify interactions of splice genes with other mutations, we performed Fisher exact test. Interestingly, each splice gene was significantly associated with one particular gene involved in epigenetic regulation. DNMT3A mutations were more often found in SF3B1mut patients than in SF3B1wt patients (35% vs 7%, P < .001; Table 2). TET2 mutations occurred more often in SRSF2mut and ZRSR2mut patients compared with SRSF2wt and ZRSR2wt patients (44% vs 22%; P = .02 and 56% vs 20%; P < .001, respectively). A trend was observed for RUNX1 mutations to be enriched in patients concomitantly mutated for SRSF2 (16% vs 6%; P = .09). Finally, ASXL1 mutations were more often seen in U2AF35mut patients than in U2AF35wt patients (45% vs 17%; P = .03).
Distribution of molecular aberrations in 154 MDS patients with at least 1 identified mutation in the 16 investigated genes.
Distribution of molecular aberrations in 154 MDS patients with at least 1 identified mutation in the 16 investigated genes.
Molecular associations of splice gene mutations (Fisher exact test)
| . | TET2mut (n = 54) . | ASXL1mut (n = 37) . | DNMT3Amut (n = 26) . | RUNX1mut (n = 16) . | TP53mut (n = 13) . | IDH1/2mut (n = 12) . | CBLmut (n = 11) . | JAK2mut (n = 9) . | EZH2mut (n = 7) . | ETV6mut (n = 2) . | NRASmut (n = 2) . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SF3B1mut (n = 37) | 9 | 4 | 13 (P < .001) | 3 | 1 | 1 | 1 | 3 | 1 | 0 | 0 |
| SRSF2mut (n = 25) | 11 (P = .02) | 6 | 4 | 4 (P = .09) | 1 | 3 | 3 | 0 | 0 | 0 | 0 |
| ZRSR2mut (n = 25) | 14 (P < .001) | 5 | 1 | 3 | 0 | 3 | 1 | 0 | 1 | 0 | 0 |
| U2AF35mut (n = 12) | 1 | 5 (P = .03) | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 |
| . | TET2mut (n = 54) . | ASXL1mut (n = 37) . | DNMT3Amut (n = 26) . | RUNX1mut (n = 16) . | TP53mut (n = 13) . | IDH1/2mut (n = 12) . | CBLmut (n = 11) . | JAK2mut (n = 9) . | EZH2mut (n = 7) . | ETV6mut (n = 2) . | NRASmut (n = 2) . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SF3B1mut (n = 37) | 9 | 4 | 13 (P < .001) | 3 | 1 | 1 | 1 | 3 | 1 | 0 | 0 |
| SRSF2mut (n = 25) | 11 (P = .02) | 6 | 4 | 4 (P = .09) | 1 | 3 | 3 | 0 | 0 | 0 | 0 |
| ZRSR2mut (n = 25) | 14 (P < .001) | 5 | 1 | 3 | 0 | 3 | 1 | 0 | 1 | 0 | 0 |
| U2AF35mut (n = 12) | 1 | 5 (P = .03) | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 |
Prognostic impact of splice gene mutations
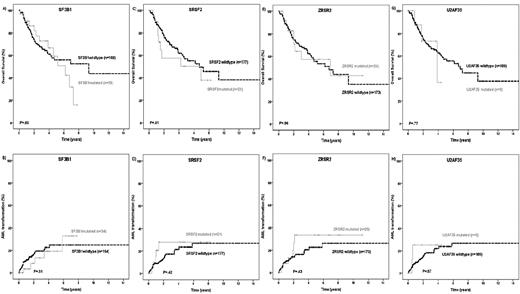
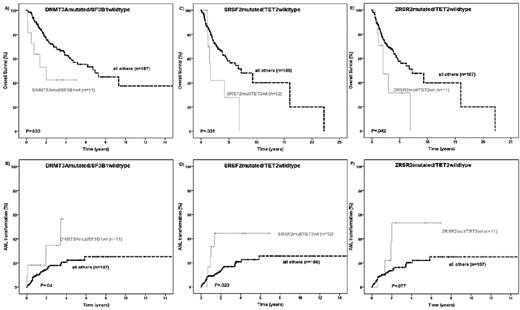
The prognostic impact of splice gene mutations was evaluated in MDS patients for whom follow-up information was available (n = 198). The median follow-up of patients alive was 31 months. In univariate analysis, OS and AML transformation rates according to the mutation status of the 4 different splice genes were similar (Figure 3A-H). Next, we evaluated these 2 endpoints in low- and high-risk MDS patients defined by their IPSS score. Except for a higher AML transformation rate of ZRSR2mut patients compared with ZRSR2wt patients in the IPSS-low or intermediate-1 subgroups (P = .022), no differences on patient outcome according to the splice gene mutation status were observed (supplemental Figures 1-2). As specific molecular associations between mutations targeting epigenetic and splice activities were identified, we analyzed the prognostic impact of these molecularly defined genotypes. No difference in patient outcome was observed when analyzing the genotypes SF3B1mut/DNMT3Amut or SF3B1mut/DNMT3Awt (data not shown). In contrast, SF3B1wt/DNMT3Amut patients had an inferior OS (hazard ratio [HR] = 2.35; 95% CI, 1.09-5.46; P = .033) and a higher risk of AML transformation (HR = 2.7; 95% CI, 1.05-7.68; P = .04; Figure 4A-B) compared with all other genotypes (SF3B1mut/DNMT3Amut or SF3B1mut/DNMT3Awt or SF3B1wt/DNMT3Awt). An inferior OS and a higher transformation rate to AML was also seen in patients defined by the genotype SRSF2mut/TET2wt in univariate analysis (OS: HR = 2.32; 95% CI, 1.05-5.11; P = .031; AML transformation: HR = 2.94; 95% CI, 1.13-7.61; P = .02; Figure 4C-D). Patients with the genotype ZRSR2mut/TET2wt had a shorter OS (HR = 2.21; 95% CI, 1.01-4.85; P = .042) and tended to have a higher rate for AML transformation (HR = 2.4; 95% CI, 0.94-6.81; P = .077; Figure 4E-F) compared with all other genotypes.
Kaplan-Meier curves for OS and time to AML transformation. (A-B) The SF3B1 mutation status. (C-D) The SRSF2 mutation status. (E-F) The ZRSR2 mutation status. (G-H) The U2AF35 mutation status in 198 MDS patients (log-rank test).
Kaplan-Meier curves for OS and time to AML transformation. (A-B) The SF3B1 mutation status. (C-D) The SRSF2 mutation status. (E-F) The ZRSR2 mutation status. (G-H) The U2AF35 mutation status in 198 MDS patients (log-rank test).
Kaplan-Meier curves for OS and time to AML transformation. (A-B) The DNMT3A/SF3B1 mutation status. (C-D) The SRSF2/TET2 mutation status. (E-F) The ZRSR2/TET2 mutation status in 198 MDS patients (log-rank test).
Kaplan-Meier curves for OS and time to AML transformation. (A-B) The DNMT3A/SF3B1 mutation status. (C-D) The SRSF2/TET2 mutation status. (E-F) The ZRSR2/TET2 mutation status in 198 MDS patients (log-rank test).
In multivariate analysis, including age, IPSS risk groups, transfusion dependence, mutation status for ASXL1, RUNX1, TP53, CBL, and the genotypes DNMT3Amut/SF3B1wt, SRSF2mut/TET2wt, and ZRSR2mut/TET2wt, the genotype ZRSR2mut/TET2wt was found to be an independent unfavorable prognostic factor for OS (HR = 3.3; 95% CI, 1.4-7.7; P = .006) and ZRSR2mut/TET2wt independently associated with a higher AML transformation rate (HR = 3.6; 95% CI, 2-4.2; P = .026; Table 3).
Univariate and multivariate analysis for OS and time to AML transformation in MDS patients
| . | OS (univariate analysis) . | OS (multivariate analysis) . | AML transformation (univariate analysis) . | AML transformation (multivariate analysis) . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | HR . | 95% CI . | P . | HR . | 95% CI . | P . | |
| Age, y (above vs below the median) | 1.9 | 1.1-3.2 | .013 | 1.7 | 0.9-3.0 | .065 | NS | |||||
| IPSS risk groups* (high vs low) | 2.2 | 1.7-2.8 | < .001 | 2.1 | 1.6-2.8 | < .001 | 2.9 | 2.1-4 | < .001 | 3.2 | 2.1-4.7 | < .001 |
| Transfusion dependence (yes vs no) | 2.9 | 1.7-5.1 | < .001 | 2.3 | 1.3-4.2 | .005 | 2.4 | 1.1-5 | .023 | 2.2 | 1-4.9 | .042 |
| ASXL1mut vs ASXL1wt | 2.0 | 1.2-3.5 | .007 | NS | 2.8 | 1.4-5.6 | .002 | NS | ||||
| RUNX1mut vs RUNX1wt | 2.0 | 1-4.1 | .044 | NS | 4.6 | 2.2-9.9 | < .001 | 3.1 | 1.3-7.4 | .009 | ||
| TP53mut vs TP53wt | 4.4 | 2.1-9.1 | < .001 | 2.0 | 1-4.7 | .041 | 3.9 | 1.5-10.2 | .006 | NS | ||
| CBLmut vs CBLwt | 2.6 | 1.1-6.0 | .027 | 4.4 | 1.7-11.6 | .002 | 4.3 | 1.7-11.3 | .003 | 4.1 | 1.4-12.4 | .01 |
| DNMT3Amut /SF3B1wt vs all other | 2.4 | 1.1-5.5 | .033 | NS | 2.7 | 1.1-7.7 | .04 | NS | ||||
| SRSF2mut /TET2wt vs all other | 2.3 | 1.1-5.1 | .031 | NS | 2.9 | 1.1-7.6 | .02 | NS | ||||
| ZRSR2mut/TET2wt vs all other | 2.2 | 1.0-4.9 | .042 | 3.3 | 1.4-7.7 | .006 | 2.4 | 0.9-6.8 | .077 | 3.6 | 2-4.2 | .026 |
| . | OS (univariate analysis) . | OS (multivariate analysis) . | AML transformation (univariate analysis) . | AML transformation (multivariate analysis) . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | HR . | 95% CI . | P . | HR . | 95% CI . | P . | |
| Age, y (above vs below the median) | 1.9 | 1.1-3.2 | .013 | 1.7 | 0.9-3.0 | .065 | NS | |||||
| IPSS risk groups* (high vs low) | 2.2 | 1.7-2.8 | < .001 | 2.1 | 1.6-2.8 | < .001 | 2.9 | 2.1-4 | < .001 | 3.2 | 2.1-4.7 | < .001 |
| Transfusion dependence (yes vs no) | 2.9 | 1.7-5.1 | < .001 | 2.3 | 1.3-4.2 | .005 | 2.4 | 1.1-5 | .023 | 2.2 | 1-4.9 | .042 |
| ASXL1mut vs ASXL1wt | 2.0 | 1.2-3.5 | .007 | NS | 2.8 | 1.4-5.6 | .002 | NS | ||||
| RUNX1mut vs RUNX1wt | 2.0 | 1-4.1 | .044 | NS | 4.6 | 2.2-9.9 | < .001 | 3.1 | 1.3-7.4 | .009 | ||
| TP53mut vs TP53wt | 4.4 | 2.1-9.1 | < .001 | 2.0 | 1-4.7 | .041 | 3.9 | 1.5-10.2 | .006 | NS | ||
| CBLmut vs CBLwt | 2.6 | 1.1-6.0 | .027 | 4.4 | 1.7-11.6 | .002 | 4.3 | 1.7-11.3 | .003 | 4.1 | 1.4-12.4 | .01 |
| DNMT3Amut /SF3B1wt vs all other | 2.4 | 1.1-5.5 | .033 | NS | 2.7 | 1.1-7.7 | .04 | NS | ||||
| SRSF2mut /TET2wt vs all other | 2.3 | 1.1-5.1 | .031 | NS | 2.9 | 1.1-7.6 | .02 | NS | ||||
| ZRSR2mut/TET2wt vs all other | 2.2 | 1.0-4.9 | .042 | 3.3 | 1.4-7.7 | .006 | 2.4 | 0.9-6.8 | .077 | 3.6 | 2-4.2 | .026 |
NS indicates not significant.
IPSS-low indicates IPSS-low risk or intermediate-1; and IPSS-high, IPSS-high or intermediate-2.
Discussion
Identification of novel targets to direct new treatment approaches remains a major challenge in MDS. Recent discovery of novel pathway mutations affecting spliceosome core components prompted us to investigate the 4 most recurrently mutated genes in a large cohort of 221 MDS patients.11 Our present study confirms the high prevalence of mutations affecting splicing activity in this heterogeneous disorder.11 Almost half of investigated patients presented a mutation in SF3B1 (16.4%), SRSF2 (11.1%), ZRSR2 (11.1%), or U2AF35 (5.4%). We identified distinct clinical phenotypes and molecular association patterns for each splice gene. While SF3B1mut patients were likely to present with RSs and reduced hemoglobin levels leading to a higher transfusion dependence rate, patients harboring SRSF2 mutations clustered in RAEB-1 and RAEB-2 subtypes and had pronounced thrombocytopenias. ZRSR2mut patients had a similar clinical phenotype, clustered in IPSS intermediate-1 and intermediate-2 risk groups, had higher bone marrow blast percentages, and often exhibited isolated neutropenias. The association between a splice gene mutation and a specific clinical phenotype remained independently significant when analyzed together with IPSS-risk group and WHO disease subtype in a linear regression model (eg, SF3B1 mutation and reduced hemoglobin levels [P = .011], or SRSF2 mutation and thrombocytopenia [P = .025]). This suggests that splice gene mutations play a causal role in the disease course. Interestingly, each splice gene mutation was associated with one concomitant mutation in a gene involved in epigenetic regulation of transcription. Although half of the patients with DNMT3A mutations also had SF3B1 mutations, TET2 mutations were significantly enriched in both SRSF2 and ZRSR2 mutated patients. Our results demonstrate U2AF35 mutations to be enriched in patients with ASXL1 mutations and confirm an earlier report that a deletion of chromosome 20 is often found in U2AF35mut patients.14 However, in contrast to Graubert et al,14 whose analysis of 150 MDS patients suggested an increased probability of secondary AML progression for patients with U2AF35 mutations, we observed no association between U2AF35 mutations and transformation to AML. Whether the suggested higher transformation rate is because of U2AF35 mutations or confounded by the presence of an ASXL1 mutation, which was consistently shown to be associated with worse OS and higher AML transformation rates in myeloid malignancies,8,10,28 must be verified in even larger cohorts of MDS patients. Of the 7 patients who carried a U2AF35 mutation and were wild-type for ASXL1, one had progression to AML and died 10 months after MDS diagnosis. It is currently debated whether the recurrent ASXL1 variation (c.1934dupG;p.Gly646TrpfsX12), which accounts for the majority of patients who we classified as mutated for ASXL1, is a bona fide somatic mutation or an artifact.29 This alteration was found in the majority of patients who we classified as mutated for ASXL1. In accordance with previous reports, we did not detect this alteration in genomic DNA from CD3+ cells of the same patients, when available.10,30,31
In contrast to some recent reports, we observed no favorable impact on OS for patients with SF3B1 mutation.16,32 This may be at least partially related to the heterogeneity of investigated cohorts: our study cohort includes a lower number of refractory anemia with RSs (RARS)/refractory cytopenia with multilineage dysplasia (RCMD)-RS or RARS-T patients compared with others (19% vs 29%)16,32 and a higher number of advanced MDS or RAEB-1 or RAEB-2 subtypes (45% vs 26%).32 However, when restricting survival analysis to patients with RSs (RARS, RCMD-RS or RARS-T; n = 36 with available clinical information), no difference on OS or AML transformation rate was observed (data not shown). These results are in accord with 2 recent reports investigating the prognostic importance of SF3B1 mutations in larger cohorts of MDS patients with RSs, which did not find an independent value within this subgroup of patients where the highest prevalence of SF3B1 mutations is found.32 We identified a negative prognostic impact on OS and higher rate of AML transformation for SF3B1wt/DNMT3Amut patients. This finding is in accord with recent reports in AML and MDS, which demonstrated a strong association of DNMT3A mutations with adverse outcome.7,33-35 Furthermore, we show that the prognostic impact of SRSF2 and ZRSR2 mutations depends on the mutation status of TET2, the gene most commonly mutated in MDS. These findings are of major interest as they link two pathways involved in the pathophysiology of MDS. Whether the identified genotype combinations are restricted to MDS or are also found in other myeloid or lymphoid malignancies needs to be established. However, the molecular crosstalk between these mutations is difficult to predict because loss of Tet2 has been demonstrated to endow cells with a growth advantage,36-38 whereas overexpression of mutated SRSF2 or U2AF35 has antiproliferative effects11 and will need to be investigated in appropriate settings. Our data support the idea that the mutations in the splice genes contribute to the clinical/biologic phenotype of the MDS clone.
In conclusion, we identified mutations of SF3B1, SRSF2, ZRSR2, and U2AF35 in 42.2% of MDS patients and found a strong association between the clinical phenotype and the different splice gene mutations. A distinct molecular pattern involving the epigenetic regulation of transcription and the splicing machinery was identified and associated with patient outcome. Although confirmation of these results in larger cohorts is necessary before redefining risk-guided therapy strategies in MDS, integration of molecular analysis of the splice genes at diagnosis may improve classification of MDS patients.
The online version of this article contains a data supplement.
Acknowledgments
The authors thank the investigators of the Groupe Francophone des Myélodysplasies (Dr M. Gardembas, Angers; Dr M. Dib, Angers; Dr C. Rose, Lille; Dr S. Cheze, Caen; Dr S. Park, Hôpital Cochin, Paris; Dr S. Natarajan-Ame, Strasbourg; Dr E. Gyan, Tours; Dr L. Legros, Nice; Dr C. Soussain, CH René Huguenin, Saint-Cloud; Dr E. Raffoux, Hôpital Saint-Louis, Paris; Pr E. Solary, Institut G Roussy, Villejuif) for including patients; L. Slama, G. Herledan, and J. Brard for technical assistance; and R. Sapena for data management of the French MDS registry.
This study was supported by the Deutsche Krebshilfe (grant 109686) and the Hannelore-Munke Fellowship (F. Damm); association Laurette Fugain (O.A.B. and F. Dreyfus); Association pour la Recherche sur le Cancer (no. 4992, 2010, D.B.); Direction de la recherche clinique AP-HP (PHRC MDS-04); and INCa génomique et fonction des gènes dans les cancers-valorisation des ressources biologiques 2008 (M.F.), INCa; and a labelization from the Ligue Nationale Contre le Cancer (O.A.B.).
Authorship
Contribution: F. Damm, O.K., O.A.B., and M.F. designed the research; F. Damm, O.K., V.G.-B., A.R., C.H.-C., V.D.V., L.C., L.S., V.C., and N.C. performed the research; A.G.-B., B.S., O.B.-R., A.S.-T., A.S.-B., F. Dreyfus, T.P., S.d.B., N.V., D.B., and C.P. contributed patient samples and clinical data; F. Damm, O.K., V.G.-B., A.R., M.A.M., N.C., and N.C.P.C. analyzed the data; F. Damm, O.K., O.A.B., and M.F. wrote the manuscript; and all authors read and agreed to the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michaela Fontenay, Service d'Hématologie Biologique, GH Broca-Cochin-Hôtel-Dieu, and Institut Cochin, 27 rue du Fg St Jacques, 75014 Paris, France; e-mail: michaela.fontenay@inserm.fr; and Olivier A. Bernard, Inserm U985, Institut Gustave Roussy, 39 rue Camille Desmoulins, 94800 Villejuif, France; e-mail: olivier.bernard@inserm.fr.