Abstract
MicroRNAs (miRNAs) are small, noncoding RNA molecules that regulate growth and differentiation. miRNAs are frequently located at cancer-specific fragile sites in the human genome, such as chromosome 7q. The nuclear oncogene SKI is up-regulated in acute myeloid leukemia (AML) with −7/del7q. Here we asked whether loss of miRNAs on chromosome 7q may explain this up-regulation. miR-29a expression was found to be down-regulated in AML with −7/del7q. Forced expression of miR-29a down-regulated Ski and its target gene, Nr-CAM, whereas miR-29a inhibition induced Ski expression. Luciferase assays validated a functional binding site for miR-29a in the 3′ untranslated region of SKI. Finally, in samples of AML patients, we observed an inverse correlation of Ski and miR-29a expression, respectively. In conclusion, up-regulation of Ski in AML with −7/del7q is caused by loss of miR-29a. miR-29a may therefore function as an important tumor suppressor in AML by restraining expression of the SKI oncogene.
Introduction
Patients with acute myeloid leukemia (AML) with monosomy 7 (−7) or deletion of 7q (del7q) have a poor prognosis. We recently reported that the nucleoprotein Ski, the cellular homologue of the avian leukosis retrovirus oncogene v-SKI, is up-regulated in AML, especially in AML with −7/del7q, which presumably represses retinoic acid–induced myeloid differentiation.1 In keeping with this, AML patients with low levels of SKI may benefit from combination therapy with retinoic acid.2 The molecular reason for up-regulation of SKI in AML with −7 is unclear. Therefore, we investigated whether a microRNA (miRNA) localized in the cancer fragile site of 7q could be a regulator of Ski. miRNAs are small RNA molecules that affect gene expression by posttranscriptional and translational mechanisms.3 Different miRNAs have been shown to regulate cell proliferation, differentiation, and apoptosis.4-6 Aberrant miRNA expression has been linked to leukemogenesis in chronic lymphocytic leukemia,7 acute lymphoblastic leukemia,8 and AML.9-11 miRNAs contribute to malignant transformation by genomic loss, because many miRNAs are tumor-suppressive and localized in cancer fragile sites.12 We describe here that expression of miR-29a localized on 7q32, is down-regulated in AML with −7/del7q and that miR-29a regulates Ski. These data suggest that loss of miR-29a may contribute to leukemogenesis by derepressing the nuclear oncogene SKI.
Methods
Patient samples, cell lines, and transfections
For detailed descriptions of patient cohorts, cell lines, and transient transfections, see supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
miRNA array
We used an oligonucleotide array (mirVANA miRNA probe set; Ambion) and tested the miRNA expression pattern of 17 AML samples. The AML study was approved by the local ethics committee, and patients and donors gave written informed consent. For details, see supplemental Methods.
Flow cytometry
For a description of flow cytometry, see Ritter et al.1
Cell proliferation assay
For details about the cell proliferation (MTT) assay, see supplemental Methods.
Western blot
For Western blot, whole-cell proteins were extracted by use of a standard protocol for RIPA lysates. After SDS-PAGE, proteins were transferred to nitrocellulose membrane and incubated with antibody anti-Ski (sc-9140; Santa Cruz Biotechnology; 1:1000) or anti–β-actin (AC-74; Sigma-Aldrich; 1:5000).
Reverse transcription, Nr-CaM PCR, and quantitative real-time PCR for miR-29a
For conditions and primers, see supplemental Methods.
SKI 3′UTR reporter constructs and luciferase gene assay
For luciferase constructs (Figure 2A) and luciferase assay, see supplemental Methods.
Statistical analysis
For scientific graphing and statistical analysis, we used GraphPad Prism 5 software (GraphPad Prism Software Inc).
Results and discussion
Microarray analysis shows miR-29a is down-regulated in AML samples with −7/del7q karyotype
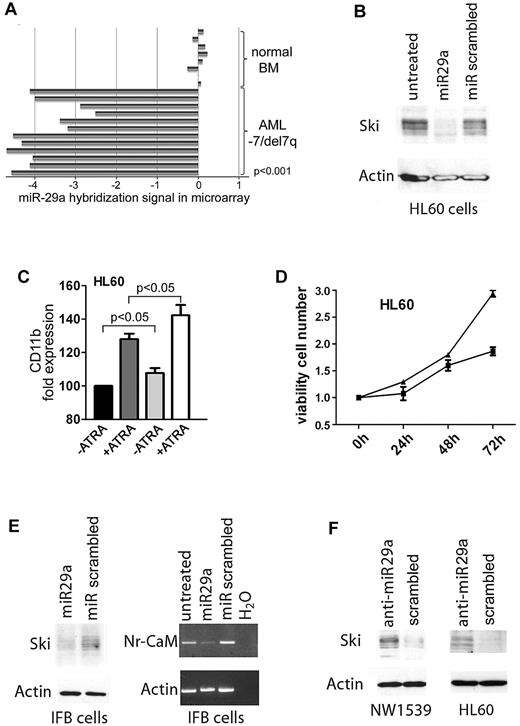
We performed an miRNA oligonucleotide array and compared the miRNA profile of normal bone marrow samples (n = 8) with that of AML samples with isolated −7/del7q or complex karyotype including −7/del7q (n = 17, cohort 1; supplemental Tables 1 and 2; blast count > 30%). A hierarchical cluster analysis demonstrated that AML samples had expression patterns different from normal bone marrow (supplemental Figure 1). Next, we looked for differential miRNA expression between normal bone marrow and AML with −7/del7q (supplemental Table 3). The only miRNA found to be down-regulated in AML samples and localized on 7q was miR-29a (Figure 1A). We then also analyzed 2 larger cohorts of patients for miR-29a expression using quantitative RT-PCR: cohort 2 (−7/del7q; n = 61; supplemental Table 4) and cohort 4 (normal karyotype; n = 54). Between these 2 cohorts, no significant different expression of miR-29a was found, which could be attributed to technical reasons; however, both cohorts showed a down-regulation compared with normal CD34-positive cells (supplemental Figure 2; supplemental Table 5). miR-29a is down-regulated in chronic lymphocytic leukemia13 and solid cancers.14,15 Because our previous results indicated an overexpression of the nuclear oncogene SKI in AML with −7/del7q,1 we examined whether miR-29a controls Ski expression.
Expression of miR-29a is down-regulated in AML with −7/del7q and regulates expression of Ski. (A) Expression of miR-29a in AML patient samples with isolated −7/del7q karyotype or complex karyotype including −7/del7q. Expression levels of miR-29a in 12 AML samples with −7/del7q as determined by microarray analysis compared with miR-29a expression in 8 samples of normal bone marrow. Five of the original 17 values for miR-29a expression in AML samples are missing. (B) miR-29a inhibits Ski expression. HL60 cells were transfected with 29a or scrambled precursor miRNA. After 24 hours, Ski expression was analyzed in whole-cell protein extracts with Western blot. Loading control was β-actin. (C) miR-29a enhances ATRA-induced differentiation of myeloid HL60 cells. Cellular differentiation of HL60 cells transfected with 29a or scrambled control precursor miRNA additionally treated with 1.5μM ATRA for 72 hours was analyzed with flow cytometry. Black bar indicates miR-scrambled; dark gray bar, miR-scrambled plus ATRA; gray bar, miR-29a; white bar, miR-29a plus ATRA. CD11b expression of miR-scrambled transfected cells was set at 100%. Results are representative of 3 independent experiments. Values shown are mean ± SEM. (D) miR-29a reduces proliferation of myeloid HL60 cells. HL60 cells were transfected with miRNA precursor 29a or scrambled control. After 0, 24, 48, and 72 hours, proliferation was tested in an MTT assay. ■ indicates miR-29a; ▴, miR scrambled. Experiment was performed 3 times in triplicate. Values shown are mean ± SEM. (E) Expression of the Ski target gene Nr-CAM is inhibited by miR-29a. IFB melanoma cells were transfected with miR-29a or scrambled precursor miRNA. After 24 hours, Ski expression was tested in whole-cell protein lysates by Western blot (left). Loading control was β-actin. Nr-CAM expression was measured by PCR (right). Control was β-actin. (F) Inhibition of miR-29a induces Ski expression. NW1539 melanoma cells (left) or HL60 cells (right) were transfected with anti–miR-29a or scrambled oligonucleotide. After 48 hours, Western blot was performed for Ski with whole-cell protein extracts. Loading control was β-actin.
Expression of miR-29a is down-regulated in AML with −7/del7q and regulates expression of Ski. (A) Expression of miR-29a in AML patient samples with isolated −7/del7q karyotype or complex karyotype including −7/del7q. Expression levels of miR-29a in 12 AML samples with −7/del7q as determined by microarray analysis compared with miR-29a expression in 8 samples of normal bone marrow. Five of the original 17 values for miR-29a expression in AML samples are missing. (B) miR-29a inhibits Ski expression. HL60 cells were transfected with 29a or scrambled precursor miRNA. After 24 hours, Ski expression was analyzed in whole-cell protein extracts with Western blot. Loading control was β-actin. (C) miR-29a enhances ATRA-induced differentiation of myeloid HL60 cells. Cellular differentiation of HL60 cells transfected with 29a or scrambled control precursor miRNA additionally treated with 1.5μM ATRA for 72 hours was analyzed with flow cytometry. Black bar indicates miR-scrambled; dark gray bar, miR-scrambled plus ATRA; gray bar, miR-29a; white bar, miR-29a plus ATRA. CD11b expression of miR-scrambled transfected cells was set at 100%. Results are representative of 3 independent experiments. Values shown are mean ± SEM. (D) miR-29a reduces proliferation of myeloid HL60 cells. HL60 cells were transfected with miRNA precursor 29a or scrambled control. After 0, 24, 48, and 72 hours, proliferation was tested in an MTT assay. ■ indicates miR-29a; ▴, miR scrambled. Experiment was performed 3 times in triplicate. Values shown are mean ± SEM. (E) Expression of the Ski target gene Nr-CAM is inhibited by miR-29a. IFB melanoma cells were transfected with miR-29a or scrambled precursor miRNA. After 24 hours, Ski expression was tested in whole-cell protein lysates by Western blot (left). Loading control was β-actin. Nr-CAM expression was measured by PCR (right). Control was β-actin. (F) Inhibition of miR-29a induces Ski expression. NW1539 melanoma cells (left) or HL60 cells (right) were transfected with anti–miR-29a or scrambled oligonucleotide. After 48 hours, Western blot was performed for Ski with whole-cell protein extracts. Loading control was β-actin.
miR-29a regulates Ski protein expression
We transfected HL60 cells with miRNA precursor 29a, 25, 183, or 335, localized on chromosome 7, and measured Ski using flow cytometry. Only miR-29a effectively lowered Ski in HL60 (supplemental Figure 3). Consistently, miR-29a reduced Ski compared with miR-scrambled in Western blot (Figure 1B). Flow cytometry revealed that overexpression of miR-29a (fold miR-29a expression compared with scrambled: 17) significantly increased expression of monocytic differentiation marker CD11b (P = .023). This could be enhanced by all-trans retinoic acid (ATRA; Figure 1C; P = .043). In keeping with this, overexpression of miR-29a decreased proliferation of HL60 cells (Figure 1D).
Ski may be a transcriptional corepressor16 or coactivator.17 In IFB melanoma cells, Ski acts as a coactivator of the neuronal cell adhesion molecule gene, Nr-CAM.18 We transfected IFB cells with miRNA precursor 29a (fold expression compared with scrambled: 25) and determined expression of Ski using Western blot and of Nr-CAM using PCR. Expression of miR-29a inhibited Ski protein and Nr-CAM gene expression, in keeping with our hypothesis (Figure 1E).
We next investigated whether inhibition of miR-29a would lead to increased Ski expression. NW1539 melanoma and HL60 cells express miR-29a (supplemental Figure 4). These cells were incubated with an inhibitor oligonucleotide against miR-29a or scrambled. Addition of anti-miR-29a induced Ski expression in NW1539 (Figure 1F) and HL60 (Figure 1F; fold down-regulation of miR-29a compared with scrambled: 10).
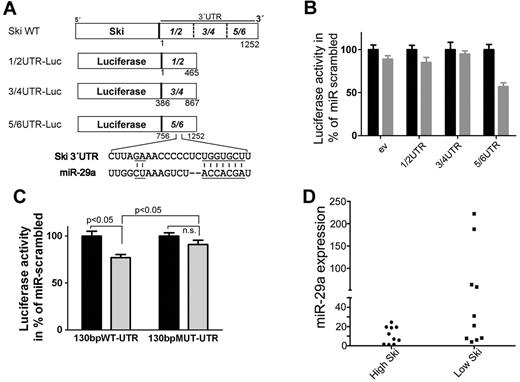
miR-29a targets the 3′UTR region of Ski
To analyze the function of miR-29a, we transfected HeLa cells with 3 different luciferase constructs of SKI wild-type 3′UTR (Figure 2A) and miRNA precursor 29a. miR-29a significantly reduced activity of the 5/6-UTR construct compared with other constructs (Figure 2B). Endogenous miR-29a also reduced activity of the 5/6-UTR construct (supplemental Figure 5), which indicates a binding site for miR-29a in this region (756-1252 bp). In silico analysis (www.targetscan.org) identified a putative binding site for miR-29a in the SKI 3′UTR at 1066-1088 bp (Figure 2A), which is conserved in Homo sapiens, mouse, rat, and dog. To confirm, we transfected HeLa cells with miRNA precursor 29a and luciferase constructs carrying wild-type or mutated predicted miR-29a binding site flanked by 130 bp of 5/6-UTR. The introduction of mutations of the putative binding region abolished the effect on luciferase activity, which supports the notion that miR-29a binds to this region (Figure 2C).
miR-29a binds to 3′UTR of SKI and is associated with low Ski expression in primary AML cells. (A) Scheme of the luciferase constructs. The 3′UTR of Ski was subdivided into 3 approximately 450-bp overlapping fragments (1/2 bp 1-465, 3/4 bp 386-867, 5/6 bp 756-1252) and cloned in the 3′direction of the luciferase gene in the pGL3 control vector. A putative binding site for miR-29a in fragment 5/6 of Ski 3′UTR at bp 1066-1088 is shown. Underlined are complementary parts of miR-29a and SKI 3′UTR. (B) miR-29a targets the 3′UTR of Ski in vitro. The empty luciferase reporter plasmid (ev) or luciferase reporter plasmids carrying the 1/2, 3/4, or 5/6 fragment of the 3′UTR of Ski (1/2-UTR, 3/4-UTR, 5/6-UTR) were cotransfected with scrambled precursor miRNA or miR-29a precursor and with pRL-TK-Luc in HeLa cells. Twenty-four hours after transfection, Firefly luciferase activity was measured. Black bars indicate miR-scrambled; gray bars, miR-29a. Results are representative of 3 individual experiments with 5 replicates. Normalized Renilla luciferase expression in cells transduced with the different SKI-3′UTR plasmids and miRna-scrambled was set at 100%. Values are mean ± SEM. (C) miR-29a targets its predicted binding site in SKI-3′UTR. The luciferase reporter plasmids 130bpWT-UTR or 130bpMUT-UTR contained a 130-bp fragment of 5/6-UTR that flanked the putative binding site in 5/6 at position 1066-1088. The original putative binding sequence CUUAGAAACCCCCUCUGGUGCCU was mutated to CUUAGCAACCTCCUCUCGUACCU. The changed bases are underlined in Figure 2A. HeLa cells were cotransfected with scrambled precursor miRNA (black bars) or miR-29a precursor (gray bars) and with pRL-TK-Luc. Twenty-four hours after transfection, Firefly luciferase activity was measured. Results are representative of 3 individual experiments with 5 replicates. Normalized Renilla luciferase expression in cells transduced with the different 3′UTR-plasmids and miRNA-scrambled was set at 100%. Values are mean ± SEM. (D) miR-29a and Ski expression are inversely associated in human AML samples. Statistical analysis of Ski Western blot data and miR-29a quantitative RT-PCR data showed a significant correlation (P = .020) of high miR-29a expression and low Ski expression in AML patient samples (n = 21).
miR-29a binds to 3′UTR of SKI and is associated with low Ski expression in primary AML cells. (A) Scheme of the luciferase constructs. The 3′UTR of Ski was subdivided into 3 approximately 450-bp overlapping fragments (1/2 bp 1-465, 3/4 bp 386-867, 5/6 bp 756-1252) and cloned in the 3′direction of the luciferase gene in the pGL3 control vector. A putative binding site for miR-29a in fragment 5/6 of Ski 3′UTR at bp 1066-1088 is shown. Underlined are complementary parts of miR-29a and SKI 3′UTR. (B) miR-29a targets the 3′UTR of Ski in vitro. The empty luciferase reporter plasmid (ev) or luciferase reporter plasmids carrying the 1/2, 3/4, or 5/6 fragment of the 3′UTR of Ski (1/2-UTR, 3/4-UTR, 5/6-UTR) were cotransfected with scrambled precursor miRNA or miR-29a precursor and with pRL-TK-Luc in HeLa cells. Twenty-four hours after transfection, Firefly luciferase activity was measured. Black bars indicate miR-scrambled; gray bars, miR-29a. Results are representative of 3 individual experiments with 5 replicates. Normalized Renilla luciferase expression in cells transduced with the different SKI-3′UTR plasmids and miRna-scrambled was set at 100%. Values are mean ± SEM. (C) miR-29a targets its predicted binding site in SKI-3′UTR. The luciferase reporter plasmids 130bpWT-UTR or 130bpMUT-UTR contained a 130-bp fragment of 5/6-UTR that flanked the putative binding site in 5/6 at position 1066-1088. The original putative binding sequence CUUAGAAACCCCCUCUGGUGCCU was mutated to CUUAGCAACCTCCUCUCGUACCU. The changed bases are underlined in Figure 2A. HeLa cells were cotransfected with scrambled precursor miRNA (black bars) or miR-29a precursor (gray bars) and with pRL-TK-Luc. Twenty-four hours after transfection, Firefly luciferase activity was measured. Results are representative of 3 individual experiments with 5 replicates. Normalized Renilla luciferase expression in cells transduced with the different 3′UTR-plasmids and miRNA-scrambled was set at 100%. Values are mean ± SEM. (D) miR-29a and Ski expression are inversely associated in human AML samples. Statistical analysis of Ski Western blot data and miR-29a quantitative RT-PCR data showed a significant correlation (P = .020) of high miR-29a expression and low Ski expression in AML patient samples (n = 21).
Expression of miR-29a and Ski is inversely correlated in human AML samples
To address whether miR-29a is negatively associated with Ski expression in vivo, we analyzed 21 AML samples with different karyotypes (cohort 3; supplemental Table 6 and supplemental Figure 6). Samples with very high miR-29a expression revealed low Ski protein expression (P = .020; Figure 2D). Consistent with robust repression of Ski by miR-29a, no single patient sample showed high-level expression of Ski in the presence of miR-29a.
In conclusion, we show here that miR-29a regulates Ski expression in AML, thus suggesting a tumor-suppressive role of miR-29a, as has been described by others.19 In contrast, Han et al20 observed that miR-29a is overexpressed and induces aberrant self-renewal in AML. We also observed a broad range of miR-29a expression in primary AML samples (Figure 2D). Because miRNAs do not regulate just one target, it is conceivable that in different AMLs, specific miRNAs may play different roles. Therefore, it is likely that the loss of miR-29a expression contributes to the up-regulation of the oncogene SKI in this subgroup of patients. This provides a mechanistic explanation for our original observation of high Ski expression in AML with −7/del7q. Because high-level expression of Ski is experimentally and clinically linked to retinoic acid resistance, this directly links drug resistance to a defined genetic aberration. Of note, it has been shown recently that miR-29a also regulates DNA methyltransferases.21 Together with our finding of miR-29a as a regulator of Ski, which is known to function in association with histone deacetylase in nuclear corepressor complexes,22 this suggests that the tumor-suppressive role of miR-29a is based at least in part on deregulated epigenetic control of leukemogenic gene expression.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the members of the AML-2003 study group for contributing patient samples and the participating patients for letting them study their tumor samples. They thank Professor Elke Jäger, Nord West Krankenhaus Frankfurt, Germany, for providing the cell line NW 1539. They thank Lukas Rycak and Lisa-Marie Weiss for technical support.
This work was supported in part by grants from the Bundesministerium für Bildung und Forschung (NGFN2: FKZ 01GS0449, NGFNplus: FKZ 01GS0880), the Deutsche Forschungsgemeinsschaft (Transregio 17, project C3; KFO 210, project 3), the Deutsche José Carreras Leukämie Stiftung, and a nonrestricted grant from Dr Reinfried Pohl.
Authorship
Contribution: S.T. designed and performed research, collected data, analyzed and interpreted data, and wrote the manuscript; T.I. performed research, collected, analyzed, and interpreted data, performed statistical analysis, and wrote parts of the manuscript; J.R. performed research experiments and analyzed data from experiments; D.O. designed research and contributed analytical tools; T.S. designed research, interpreted data, and wrote parts of the manuscript; and A.N. designed research, interpreted and analyzed data, performed statistical analysis, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Andreas Neubauer, Department of Hematology, Oncology and Immunology, Philipps University of Marburg, Medical Center of the University Giessen and Marburg, Baldingerstrasse, 35033 Marburg, Germany; e-mail: neubauer@ailer.ni-marburg.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal