Abstract
Clonal evolution of the leukemogenic compartment may contribute to alter the therapeutic response in acute lymphoblastic leukemia (ALL). Using xenotransplantation of primary leukemia cells, we evaluated the phenotypic and genetic composition of de novo resistant very high risk precursor B-cell ALL, a subgroup defined by the persistence of minimal residual disease despite intensive chemotherapy. Analysis of copy number alterations (CNAs) showed that the xenografted leukemia, even when reconstituted from 100 cells, remained highly related to the diagnostic sample, with minor changes in CNAs, mostly deletions, emerging in most cases in the first passage into mice. At the single-cell level, the pattern of monoallelic and biallelic deletions of the CDKN2A locus revealed distinct leukemia subpopulations, which were reproducibly tracked in xenografts. In most very high risk ALL cases, the predominant diagnostic clones were reconstituted in xenografts, as shown by multiplex polymerase chain reaction analysis of immunoglobulin and T-cell receptor loci. In other cases, the pattern in CNAs and immunoglobulin and T-cell receptor rearrangement was less concordant in xenografts, suggesting the outgrowth of subclones. These results unequivocally demonstrate the existence of clonally closely related but distinct subsets of leukemia initiating cells in ALL, which has important implications for drug development and preclinical disease modeling.
Introduction
For children with precursor B acute lymphoblastic leukemia (ALL) that relapse on current intensive chemotherapy regimens, second-line therapy is challenging.1 Diagnostic markers that are predictive of poor treatment response are scarce, and modern treatment protocols base risk stratification mostly on in vivo response to treatment by monitoring persistence of minimal residual disease (MRD) after induction chemotherapy.2 Using xenotransplantation in mice, which provides new opportunities for functional investigation in ALL,3-7 we established a preclinical model of a subgroup of patients with very high risk for relapse (VHR-ALL),8 as identified by MRD.9,10
The genetic basis of resistant disease is ill defined. Single nucleotide polymorphism (SNP) array analyses revealed that the number of focal copy number alterations (CNAs) is limited to 6 to 8 CNAs per case in ALL.11 Studies in twins provide insight in the possible sequence of events that are associated with progression to overt disease from a common ancestral clone.7,12 The most frequent CNAs in ALL involve master transcription factors regulating lymphoid development, such as Ikaros family and tumor suppressor genes of the INK4A/CDKN2A/B locus that may represent tumor driving events. Surprisingly, only a limited number of additional changes were detected at relapse, often related to a minor subpopulation in the matched diagnostic sample.13,14 A candidate gene sequencing approach has identified mutations in several genes in relapsed ALL, mostly detectable in matched diagnosis samples already, sometimes only in minor subpopulations.15 Thus, leukemia progression does not appear to be associated with a high degree of ongoing genetic instability in ALL but rather with clonal evolution and selection of a limited number of genetic lesions.
Several reports based on xenotransplantation of ALL in NOD/SCID mice have led to the hypothesis that ALL may be maintained from a rare subpopulation of leukemia initiating cells (LICs).4,16 This concept has been challenged by observations in syngeneic mouse models that indicated a much higher frequency of ALL cells with repopulation capacity17-19 and by the observation that both sorted leukemia cells from immature (CD19−/CD34+) and from mature (CD19+/CD34+) ALL compartments exhibited comparable leukemia repopulating capacity in NOD/scidIL2Rγnull (NSG) mice.5,18,19 Furthermore, 2 most recent studies provide clear evidence for a dynamic pattern of clonal diversity of the LIC compartment in TEL/AML1-positive ALL20 and BCR/ABL-positive leukemias.21 Clonal evolution of the leukemogenic compartment may contribute to disease progression and resistance to therapy.
Here we provide important new functional and genetic data to support the notion that the LIC in ALL consists of highly related subclones that evolve with distinctive additional genetic lesions and that the clonal composition of bulk VHR-ALL samples can be stably propagated by xenotransplantation to constitute a renewable source of leukemia cells, closely related to the original sample. Our results justify the use of unsorted samples to model disease in this system and provide new avenues to investigate the consequences of therapy on the composition of the LIC compartment and disease progression.
Methods
Details can be found in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Patient samples
ALL cells were recovered from cryopreserved anonymized bone marrow aspirates from patients enrolled in the ongoing ALL-BFM 2000 study and had given informed consent in accordance with the Declaration of Helsinki. Approval was obtained from the Institutional Review Board of the Medical School Hannover and the local Institutional Review Board for all participating centers in the trial ALL-BFM 2000. Samples were anonymized with labels that refer to the clinical risk stratification. Depending on the MRD result during induction therapy (MRD1 + 2) and during consolidation therapy (MRD3), patients are defined here as: standard risk (SR) if MRD1 + 2 was negative, high risk (HR) if MRD1 + 2 was positive less than or equal to 10−3, and VHR if HR patients were still positive for MRD3 (supplemental Table 1). Cases are anonymized with a label according to the risk group and with a unique number.
Xenograft model
Primary ALL cells were recovered from cryopreserved presentation samples by scraping the required amount of cells with a sterile scalpel from the cryotube kept on dry ice, washed in phosphate-buffered saline, and transplanted intrafemorally into NSG mice. Unless stated otherwise, 1 million viable cells were transplanted (as assessed by trypan blue exclusion, supplemental Table 2). Engraftment was followed every 3 to 4 weeks (supplemental Table 2) by flow cytometry in the peripheral blood of transplanted animals, using mouse-specific, anti-CD45 combined with human-specific anti-CD45 and anti-CD19 antibodies. Xenografts are labeled with identifiers corresponding to the anonymized patient sample. This labeling system is used consistently for publication. For each xenograft sample, information about the transplantation and identification of the mouse is recorded in our database for later reference.
Flow cytometry
Immunophenotype analyses of primary ALL samples at diagnosis and of xenograft samples were performed as described.22
Affymetrix Genome-Wide Human SNP Version 6.0 microarrays
DNA from paired primary leukemic and remission samples as well as matching DNA from xenografts were hybridized on Affymetrix Genome-Wide Human SNP Version 6.0 microarrays according to the manufacturer's protocols (performed by Atlas Biolabs).
Raw intensity data were analyzed using Affymetrix Genotyping Console for quality control and generation of genotype calls. Paired DNA copy number and paired loss of heterozygosity (LOH) were analyzed using the Partek Genomics Suite software (Partek Incorporated).
Fluorescence in situ hybridization
Cryopreserved cells were thawed and fixed with methanol/acetic acid (3:1). A total of 2000 cells were dropped per slide for fluorescence in situ hybridization (FISH) with the LSI CDKN2A Spectrum Orange/CEP9 Spectrum Green FISH Probe Kit (Vysis/Abbott). The LSI CDKN2A probe hybridizes to the 9p21 locus, including CDKN2A/B. Slides were hybridized and evaluated as described.23
Analysis of clonal Ig/TCR rearrangements
Detection of patient-specific junctional regions of Ig and TCR genes was performed following the protocol established for the clinical follow-up analyses of MRD, which has been established and validated by an international study group and is reported in several publications (supplemental Table 3), summarized by van der Velden et al.24
Results
Reconstitution of de novo resistant ALL is faster than SR ALL in NSG mice
To model the relevant risk groups that emerge from in vivo response to treatment,9 samples from 17 cases with de novo resistant VHR disease (VHR-ALL by MRD) and from 15 cases with SR ALL (SR-ALL, control group) were selected from the repository of the ALL-BFM-2000 study25 and transplanted into NSG mice (supplemental Table 1), extending our initial series.8 We also included 4 cases with HR ALL. Using orthotopic intrafemoral transplantation without conditioning, engraftment rates of > 70% were observed. A total of 1 million viable cells, as determined by trypan blue staining and microscopy, were injected unless stated otherwise at first passage in NSG mice (supplemental Table 2). In 3 cases, less cells were available (0.3-0.7 million). In these cases, engraftment was not delayed compared with the median time of the respective risk group. Subsequent passages were performed consistently with injection of 1 million cells. Reconstitution of the leukemia occurred faster in mice transplanted with VHR-ALL cases (median of > 75% of patients engrafted at 8 weeks) than in animals transplanted with SR-ALL cells (median of > 75% of patients engrafted at 20 weeks, P < .05; Figure 1A). All leukemias that were retransplanted (24 of 28 samples, supplemental Table 2) repopulated secondary recipient animals with high engraftment levels in bone marrow and spleen, demonstrating conserved NSG repopulating capacity. Strikingly, the time to engraftment was decreased in the second passage in NSG mice for most cases with SR-ALL (median engraftment time, 13.25 weeks for primary transplants vs 6.5 weeks for secondary transplants, P < .01), whereas it was unchanged for VHR-ALL cases, suggesting that leukemia cells from de novo drug-resistant cases are more adapted to overcome the xenograft barrier compared with SR-ALL (Figure 1A-B). Xenografted VHR-ALL cells were serially passaged up to 6 times in mice with reconstitution of the full phenotype in mice (Figure 1B; supplemental Table 2). Very high yields of ALL cells were recovered from spleens (Figure 1C). Clinical symptoms of central nervous system disease (eg, palsy or exophthalmoses) were not detected (data not shown). As supported also by others,3,5,18 these data indicate that xenotransplantation can serve to amplify normally rare primary ALL cells and as model to study resistant disease, provided the consequences of selective pressure from the xenograft barrier are better understood.
Engraftment characteristics over serial transplantation of primary ALL cells in NSG mice. Cryopreserved diagnostic material was thawed, and 1 million cells were transplanted by intrafemoral injection into NSG mice 6 to 10 weeks of age. Mice were checked every 4 weeks by flow cytometry for human engraftment. Positive engraftment was defined as > 1% human CD19/CD45-positive and mouse CD45-negative cells in the peripheral blood. (A) Analysis of the time to engraftment for 9 VHR-ALL (black) and 13 SR-ALL (gray) patients. The time to engraftment in weeks is plotted for primary (solid lines) and secondary (dashed lines) transplantations. Statistical evaluation was performed with the log-rank test and indicates significant differences for the time to engraftment between VHR-ALL and SR-ALL (P < .05) and between primary and secondary transplantation for SR-ALL (P < .01). (B) Time to engraftment was analyzed for 7 VHR-ALL patients with up to 6 rounds of serial passages. (C) Number of cells harvested from spleens per mouse at death, when the percentage of human cells in the peripheral blood reached 70%. The mean value was 500 million for VHR-ALL and 650 million cells for SR-ALL xenografts (indicated as horizontal line).
Engraftment characteristics over serial transplantation of primary ALL cells in NSG mice. Cryopreserved diagnostic material was thawed, and 1 million cells were transplanted by intrafemoral injection into NSG mice 6 to 10 weeks of age. Mice were checked every 4 weeks by flow cytometry for human engraftment. Positive engraftment was defined as > 1% human CD19/CD45-positive and mouse CD45-negative cells in the peripheral blood. (A) Analysis of the time to engraftment for 9 VHR-ALL (black) and 13 SR-ALL (gray) patients. The time to engraftment in weeks is plotted for primary (solid lines) and secondary (dashed lines) transplantations. Statistical evaluation was performed with the log-rank test and indicates significant differences for the time to engraftment between VHR-ALL and SR-ALL (P < .05) and between primary and secondary transplantation for SR-ALL (P < .01). (B) Time to engraftment was analyzed for 7 VHR-ALL patients with up to 6 rounds of serial passages. (C) Number of cells harvested from spleens per mouse at death, when the percentage of human cells in the peripheral blood reached 70%. The mean value was 500 million for VHR-ALL and 650 million cells for SR-ALL xenografts (indicated as horizontal line).
ALL xenografts retain phenotypic properties of the diagnostic samples
Alteration of the microenvironment has been shown to influence the leukemia phenotype in a model of MLL rearranged leukemia.26 Because passage in NSG mice could influence the pattern of leukemia-associated cell surface markers with respect to maturation and differentiation stage, we compared the immunophenotype of xenografts after successive passages in NSG mice to the diagnostic sample using the same panel of antibodies for 5 cases for which sufficient material was available (Figure 2; supplemental Table 4A). In addition, we compared the immunophenotypes of 11 additional cases from xenografts to the diagnostic data from the reference laboratory of the BFM study (supplemental Table 4B). In general, the expression of the lineage-associated cell surface markers CD45, CD19, CD10, CD20, CD22, CD38, and CD34, and the pattern of CD7, CD13, CD33, and CD117 coexpression were very similar. Discrepancies were detected in single xenografts in some cases for the markers CD33 (6 of 16), CD20 (3 of 16), CD34 (3 of 16), CD22 (3 of 15), CD13 (2 of 15), CD117 (1 of 14), CD7 (1 of 15), and CD45 (1 of 16). We may slightly overestimate discrepancies in the 11 cases where direct comparison based on the same antibody panel was not available. These changes may reflect minor differences after selection in NSG mice in the context of a xenogenic environment. Based on these observations, it appears justified to continue to immunophenotype xenografts as part of the validation of this preclinical model system.
Primary human ALL cells are phenotypically stable in serial xenotransplantation. The immunophenotype of 2 representative cases was determined in the same experiment on cryopreserved diagnostic cells and cells from multiple serial passage in NSG mice. Xenografts from 5 passages were analyzed for case VHR-03 (A) and 2 passages for case SR-09 (B). At least 10 000 viable cells were gated based on forward-side scatter properties, and histograms representing number of events versus signal intensity are shown.
Primary human ALL cells are phenotypically stable in serial xenotransplantation. The immunophenotype of 2 representative cases was determined in the same experiment on cryopreserved diagnostic cells and cells from multiple serial passage in NSG mice. Xenografts from 5 passages were analyzed for case VHR-03 (A) and 2 passages for case SR-09 (B). At least 10 000 viable cells were gated based on forward-side scatter properties, and histograms representing number of events versus signal intensity are shown.
Because VHR-ALL patients are identified based on persistence of significant MRD loads in vivo and given an earlier study correlating in vitro drug resistance patterns to outcome for ALL,27 we hypothesized that a major phenotypic feature of VHR-ALL would be resistance to multiple chemotherapeutic agents in vitro. To better reflect the protective bone marrow niche, which contributes to drug resistance in different models,28,29 we assayed drug response profiles of xenograft cells cocultured on immortalized human mesenchymal stroma cells.8,30 Xenografts of SR-ALL cells were sensitive to chemotherapeutic agents used for induction chemotherapy (daunorubicine, vincristine and cytarabine), with one exception (SR-02 was not responsive to cytarabine). In contrast, VHR-ALL cells were at least 10-fold less sensitive to these 4 drugs, and resistance to dexamethasone correlated with poor response to prednisone in vivo (Figure 3; supplemental Table 2).8 Drug response profiles remained mostly unchanged over up to 2 passages in NSG mice for SR-ALL cells and up to 4 passages for VHR-ALL cells (Figure 3; supplemental Table 2). These results extend findings from our earlier study,8 where some of these samples were used to test for obatoclax-induced steroid resensitization of VHR-ALL samples. In addition, we show that the drug response profiles remain stable over serial transplantation. Our results indicate that an adaptation to the mouse environment rather than a selection of ALL cells that are intrinsically more resistant to chemotherapeutic agents might be responsible for the acceleration of engraftment in cells from SR patients.
Drug response profiles of xenografts remain stable. Xenograft cells from up to 4 passages from 3 VHR-ALL (A) and 4 SR-ALL (B) were cocultured on hTERT-immortalized mesenchymal stroma cells and treated for 3 days with increasing concentrations of chemotherapeutic agents. Drug concentrations (in nanomolar) are given on the x-axis in a log scale. Measurement of ALL cell survival using 7-AAD and flow cytometry and compared with vehicle-treated controls revealed conserved resistance profiles. DEX indicates dexamethasone; VCR, vincristine; DNR, daunorubicin; and AraC, cytarabine. Experiments were performed twice, and one representative example is shown.
Drug response profiles of xenografts remain stable. Xenograft cells from up to 4 passages from 3 VHR-ALL (A) and 4 SR-ALL (B) were cocultured on hTERT-immortalized mesenchymal stroma cells and treated for 3 days with increasing concentrations of chemotherapeutic agents. Drug concentrations (in nanomolar) are given on the x-axis in a log scale. Measurement of ALL cell survival using 7-AAD and flow cytometry and compared with vehicle-treated controls revealed conserved resistance profiles. DEX indicates dexamethasone; VCR, vincristine; DNR, daunorubicin; and AraC, cytarabine. Experiments were performed twice, and one representative example is shown.
LICs are frequent in xenografts of VHR-ALL samples
For many applications of the ALL xenograft model, it will be important to better understand the hierarchy in the malignant hematopoietic tissue with respect to its LIC compartment. Indeed, in analogy to other forms of leukemia,31 it is conceivable that a restricted compartment of LIC could constitute a reservoir of resistant cells. To functionally evaluate the LIC compartment, we performed serial dilution experiments by intrafemoral injection of ALL cells in NSG mice. In 4 of 6 VHR-ALL cases, 100 xenograft cells were sufficient to fully reconstitute the leukemia without conditioning the mice (Table 1; supplemental Table 5). Flow cytometry analyses of the leukemia-associated cell surface markers revealed complete conservation of the immunophenotype of the leukemia cells resulting from transplantation with 100 cells (supplemental Figure 1). Our results are in line with similar observations by others for precursor B-ALL32 and T-ALL.33 We included samples from SR-ALL patients and cryopreserved primary ALL cells whenever available. Overall, 10 000 xenograft VHR-ALL cells were enough to repopulate leukemia in all cases, despite the xenograft barrier (Table 1; supplemental Table 5). Together with other reports,32-34 our data suggest that the LIC frequency could be higher than initially anticipated in ALL.
Serial dilution of 4 VHR-ALL xenografts by intrafemoral and intravenous transplantation
| Patient . | Round of transplantation . | Cell dilution . | Intrafemoral engrafted/total . | Intravenous engrafted/total . | Mean engraftment in bone marrow, % . |
|---|---|---|---|---|---|
| VHR-01 | Second | 1 000 000 | 2/4 | 3/3 | 88 |
| 10 000 | 3/3 | 3/3* | 86 | ||
| 100* | 2/3* | 0/3 | 96 | ||
| VHR-02 | Third | 1 000 000 | 1/2 | 2/3 | 94 |
| 10 000 | 3/3 | 1/3* | 97 | ||
| 100* | 2/3* | 0/3 | 90 | ||
| VHR-03 | Third | 1 000 000 | 2/2 | 2/2 | 94 |
| 10 000 | 2/2 | 2/2* | 97 | ||
| 100* | 1/2* | 0/2 | 77 | ||
| VHR-04 | Second | 1 000 000 | 2/2 | 2/3 | 99 |
| 10 000 | 1/3 | 2/3* | 99 | ||
| 100* | 1/3* | 0/3 | 99 |
| Patient . | Round of transplantation . | Cell dilution . | Intrafemoral engrafted/total . | Intravenous engrafted/total . | Mean engraftment in bone marrow, % . |
|---|---|---|---|---|---|
| VHR-01 | Second | 1 000 000 | 2/4 | 3/3 | 88 |
| 10 000 | 3/3 | 3/3* | 86 | ||
| 100* | 2/3* | 0/3 | 96 | ||
| VHR-02 | Third | 1 000 000 | 1/2 | 2/3 | 94 |
| 10 000 | 3/3 | 1/3* | 97 | ||
| 100* | 2/3* | 0/3 | 90 | ||
| VHR-03 | Third | 1 000 000 | 2/2 | 2/2 | 94 |
| 10 000 | 2/2 | 2/2* | 97 | ||
| 100* | 1/2* | 0/2 | 77 | ||
| VHR-04 | Second | 1 000 000 | 2/2 | 2/3 | 99 |
| 10 000 | 1/3 | 2/3* | 99 | ||
| 100* | 1/3* | 0/3 | 99 |
Four VHR-ALL patients from different amplification rounds were transplanted intrafemorally and intravenously. Engraftment was considered positive when 1% of human CD19/CD45+ mouse CD45− cells were detected in peripheral blood by flow cytometry, and spleens and bone marrows were engrafted at harvest.
Lowest dilution that resulted in engraftment.
Xenografted ALL cells are clonally highly related to the corresponding diagnostic sample
Given the occurrence of multiple genetic aberrations in ALL and the evidence for a prenatal origin for certain genetic subtypes of ALL with the acquisition of additional CNAs, even before the leukemia becomes clinically apparent,7 it is possible that leukemia samples will be constituted by a pool of related subclones. Extensive studies have established that in most ALL cases a limited number of in part recurrent CNAs can be identified.11,14 Concordance of genetic aberrations detected by SNP analysis in xenograft samples compared with diagnostic samples was suggested to correlate to some extent with poor survival in BCR/ABL-positive ALL.21 To get insight into the genetic stability of xenografts on serial passage in NSG mice, we performed genome-wide copy number and LOH analysis using Affymetrix SNP Version 6.0 arrays. We compared diagnostic and remission samples of 6 ALL patients with the corresponding samples after serial transplantation in NSG mice (Figure 4; supplemental Table 6). As expected, a low number of focal CNAs (> 100 < 1000 kb) and whole arm or total chromosome abnormalities were detected in the diagnostic samples, most of them previously reported in larger studies. In all cases, CNAs corresponding to expected Ig and TCR gene rearrangements were stably detectable over serial transplantation. Only in one case (VHR-04), an additional CNA corresponding to an incomplete rearrangement of the Ig λ (IgL) occurred (Table 2; supplemental Table 6).
Genetic lesions remain stable in xenograft cells. A copy number heat map was produced from log2 ratio Affymetrix SNP Version 6.0 analyses of 6 ALL patients and their corresponding xenografts of up to 3 subsequent amplification rounds. All CNAs of > 100 kb in size were included, also showing Ig/TCR gene rearrangements. Dx indicates diagnostic sample; and 1st, 2nd, and 3rd indicates first, second, and third amplification round, respectively. *The xenografts were obtained from transplantations with 100 cells.
Genetic lesions remain stable in xenograft cells. A copy number heat map was produced from log2 ratio Affymetrix SNP Version 6.0 analyses of 6 ALL patients and their corresponding xenografts of up to 3 subsequent amplification rounds. All CNAs of > 100 kb in size were included, also showing Ig/TCR gene rearrangements. Dx indicates diagnostic sample; and 1st, 2nd, and 3rd indicates first, second, and third amplification round, respectively. *The xenografts were obtained from transplantations with 100 cells.
Changes in CNA between diagnosis and xenograft
| Sample . | Gain or loss of CNA . | Cytoband . | Copy number . | Overlapping genes . | Overlapping miRNAs . |
|---|---|---|---|---|---|
| Lesions detected only in xenografts, not in matched diagnostic samples | |||||
| VHR-01 | Gain | 9p | Deletion | Many | 7 |
| Gain | 12p13 | cnLOH | Many, including ETV6 | 4 | |
| Gain | 22q | Amplification | Many | 26 | |
| VHR-02 | Gain | 9p21.3 | Deletion | C9orf53, CDKN2A, CDKN2BAS, CDKN2B Intron of ZCCHC7 | None |
| Gain | 9p13.2 | Deletion | None | ||
| VHR-03 | Gain | 7q22.3 | Deletion | PIK3CG | None |
| VHR-04 | Gain | 22q11.22 | Deletion | VPREB1, IgL | hsa-miR649 |
| SR-02 | Gain | 1p36.13 | Amplification | CROCCL1, ESPNP, MST1P2, MST1P9, NBPF1 | None |
| Gain | 1q23.3 | Deletion | None | ||
| Gain | 3q22.1 | Amplification | F11R, ITLN2 ALG1L2, LOC729375 | None | |
| HR-03 | Gain | 9q34.11; 22q11.23 | Amplification | BCR-ABL1, many | 13 |
| Gain | 1q | Amplification | Many | 45 | |
| Lesions not detected in xenografts, only in matched diagnostic samples | |||||
| VHR-02 | Loss | 4q31.3 | Deletion | 20.34% of FBXW7 | hsa-miR3140 |
| VHR-03 | Loss of CNA in subclone | 1q42.11-1q42.13 | Deletion | Many | 3 |
| Loss of CNA in subclone | 9p | Deletion | Many | 7 | |
| Loss of CNA in subclone | 14q12 | Deletion | AP4S1, C14orf126, COCH, HEATR5A, HECTD1, MIR624, STRN3 | None | |
| SR-02 | Loss | 7p21.3 | Amplification | None | None |
| HR-03 | Loss of CNA in subclone | 9p | Amplification | Many | 7 |
| Sample . | Gain or loss of CNA . | Cytoband . | Copy number . | Overlapping genes . | Overlapping miRNAs . |
|---|---|---|---|---|---|
| Lesions detected only in xenografts, not in matched diagnostic samples | |||||
| VHR-01 | Gain | 9p | Deletion | Many | 7 |
| Gain | 12p13 | cnLOH | Many, including ETV6 | 4 | |
| Gain | 22q | Amplification | Many | 26 | |
| VHR-02 | Gain | 9p21.3 | Deletion | C9orf53, CDKN2A, CDKN2BAS, CDKN2B Intron of ZCCHC7 | None |
| Gain | 9p13.2 | Deletion | None | ||
| VHR-03 | Gain | 7q22.3 | Deletion | PIK3CG | None |
| VHR-04 | Gain | 22q11.22 | Deletion | VPREB1, IgL | hsa-miR649 |
| SR-02 | Gain | 1p36.13 | Amplification | CROCCL1, ESPNP, MST1P2, MST1P9, NBPF1 | None |
| Gain | 1q23.3 | Deletion | None | ||
| Gain | 3q22.1 | Amplification | F11R, ITLN2 ALG1L2, LOC729375 | None | |
| HR-03 | Gain | 9q34.11; 22q11.23 | Amplification | BCR-ABL1, many | 13 |
| Gain | 1q | Amplification | Many | 45 | |
| Lesions not detected in xenografts, only in matched diagnostic samples | |||||
| VHR-02 | Loss | 4q31.3 | Deletion | 20.34% of FBXW7 | hsa-miR3140 |
| VHR-03 | Loss of CNA in subclone | 1q42.11-1q42.13 | Deletion | Many | 3 |
| Loss of CNA in subclone | 9p | Deletion | Many | 7 | |
| Loss of CNA in subclone | 14q12 | Deletion | AP4S1, C14orf126, COCH, HEATR5A, HECTD1, MIR624, STRN3 | None | |
| SR-02 | Loss | 7p21.3 | Amplification | None | None |
| HR-03 | Loss of CNA in subclone | 9p | Amplification | Many | 7 |
CNAs that changed from diagnosis to xenograft material (additional/lost deletions, amplifications, and cnLOH) are presented. If > 10 genes or 3 miRNAs were in one CNA, it was noted as many or the number was listed.
We found up to 8 CNAs (deletions and amplifications) and copy neutral LOHs (cnLOH) ranging in size from single gene aberrations to chromosome whole arm deletions/amplifications at diagnosis, with the exception of one high-hyperdiploid patient who had 30 whole chromosome amplifications. On passage into NSG mice, all but 6 lesions found in the diagnostic samples were maintained (Table 2). In one case (VHR-02), a deletion of the FBXW7 gene at 4q31.3 was lost after transplantation; whereas in VHR-03, deletions at 1q42, the whole 9p arm, and 14q12, which were detected in the diagnostic sample, were not found in corresponding xenografts. Analysis of copy number ratios indicated that subclones were present in the diagnostic samples harboring these deletions, which were lost in the first round of transplantation. Two amplifications, one in sample SR-02 that involves 7p21 with no known gene detected in the diagnostic sample and one in sample HR-03 at 9p, present in a subclone at diagnosis, were not detected in xenografts.
In addition to that, only few lesions occurred in xenograft samples that were not detected in the corresponding diagnostic sample, namely, up to 2 deletions and up to 2 amplifications per sample (Table 2; Figure 4; supplemental Table 6). Deletions at 9p loci are frequent in childhood ALL11,35,36 and proposed to be associated with poor risk. In 3 of the 4 analyzed VHR cases, additional deletions detected in xenograft but not in diagnostic samples occurred at 9p, involving genes that control B-cell development, such as CDKN2A and CDKN2B. Xenografts of VHR-01 had a deletion of the whole 9p chromosome arm; whereas in VHR-02, 2 focal deletions only deleted CDKN2A and B, along with the C9orf53-gene at 9p21.3, and the ZCCHC7 gene at 9p13.2, respectively. Last, a homozygous deletion at 9p21 and LOH over the whole 9p arm was present in xenografts of VHR-03 also present as subclone in the diagnostic sample. However, a complete deletion of the whole 9p arm was detected in a fraction of the cells at diagnosis as suggested by the analysis of copy number ratios (Table 2; Figure 4; supplemental Table 6).
Other CNAs emerging in xenografts were detected for VHR-01 at 12p13 (cnLOH, including ETV6) and at 22q (whole arm amplification), for VHR-03 at 7q22 deleting the gene for PIK3CG, for VHR-04 at 22q11 the locus of the IgL rearrangement including VPREB1, and for SR-02 at 1q23 (genes for F11R and ITLN2). In xenografts of SR-02, amplifications at 3q22 (ALG1L2) and 1p36 (genes for CROCCL1, ESPNP, MST1P2, MST1P9, and NBPF1) were also found. Interestingly, xenografts that were derived from an HR patient with BCR-ABL positive and high-hyperdiploid leukemia were remarkably stable but showed an additional amplification of the BCR-ABL fusion gene. With the exception of the abnormalities at 20p and 14q12, these have been described11,14 to occur in ALL samples, suggesting that these abnormalities are not selected for randomly in NSG mice (Table 2; Figure 4; supplemental Table 6).
To evaluate the genetic stability from passage to passage in the mice, we looked at cells from 2 passages in duplicates and transplanted at limiting dilution with only 100 cells. Interestingly, we observed that differences between diagnostic and xenografted material were present in all mice of all passages and in serial dilution cases with the exception of 2 mice from VHR-03 where the deletion at 7q22 was not detectable (Figure 4; supplemental Table 6). Taken together, these observations suggested that clonal selection may occur to some degree, mostly during the first xenotransplantation already, but that overall the xenograft samples are highly related to the original diagnostic samples.
Persistence of dominant subclones with deletions at the CDKN2A/B locus in xenografts
Given the frequent occurrence of CNAs on chromosome 9p and evidence for the acquisition of additional lesions in this region in xenografts, we performed FISH to assess the clonal heterogeneity with respect to deletions in 9p21 (including the gene for CDKN2A and CDKN2B) at the single-cell level. We detected 9p21 deletions in subclones in most of the xenograft cases, and these subclones were predominant in 10 of 24 cases (supplemental Table 7). To assess differences between primary samples and xenograft cells, we included matched diagnostic samples from 9 ALL cases, including those from which SNP data were available (Figure 5; supplemental Table 7) and those that showed 9p21 deletions in the xenograft sample. As expected, we detected the existence of subpopulations with respect to their heterozygous or homozygous 9p21 deletion status. In all but one case, the major subclone was maintained in xenografts, although with relative changes in the distribution of respective subclones in some instances. As exception, xenografts of VHR-01 selected for heterozygous deletion of 9p, which was present as minor subclone at diagnosis. In 3 samples (VHR-01, SR-09, and SR-11) clones with heterozygous 9p21 deletions became predominant, indicating selection for heterozygous events. In addition, SR-11 xenografts further showed a gradual shift toward heterozygous and homozygous clones on serial transplantation (Figure 5). The existence of subclones with differences at the 9p21 locus have been described in cytogenetic reports but, to our knowledge, has not been evaluated carefully with respect to its importance in the evaluation of the clonal compartment in ALL.23,37 Our data show that xenotransplantation results in only limited changes overall; however, shifts in the distribution of subclones with respect to particular lesions do occur, indicating that selection of preexisting subclones does occur.
Interphase FISH analysis of 9p21 (CDKN2A) lesions reveals the existence of genetically distinct subclones. The clonal composition of ALL samples was analyzed using dual-color FISH probes against 9p21 (CDKN2A, red) and the centromer of chromosome 9 (CEN9, green) as control in samples from diagnosis and corresponding xenografts. (A) Representative images are shown, indicating wild-type cells with no deletion in the 9p21 locus (left), a heterozygous deletion (middle), and a homozygous deletion (right). (B) The clonal distribution was analyzed in samples from up to 6 rounds of serial transplantation. A total of 200 cells were counted in each sample. The original data are reported in supplemental Table 7. ALL cells with germline, heterozygous deletion, and homozygous deletions are represented by colored dots of size proportional to the percentage of cells in each category in relation to the total cell number as indicated in the legend. One case, VHR-01, shows clonal selection toward additional deletions at this locus, whereas the 4 other cases analyzed are more stable with respect to the clonal composition based on this genetic feature. Original magnifications ×40 (Leica LX Widefield Microscope DMI6000B with the DFC 350 FX R2 camera).
Interphase FISH analysis of 9p21 (CDKN2A) lesions reveals the existence of genetically distinct subclones. The clonal composition of ALL samples was analyzed using dual-color FISH probes against 9p21 (CDKN2A, red) and the centromer of chromosome 9 (CEN9, green) as control in samples from diagnosis and corresponding xenografts. (A) Representative images are shown, indicating wild-type cells with no deletion in the 9p21 locus (left), a heterozygous deletion (middle), and a homozygous deletion (right). (B) The clonal distribution was analyzed in samples from up to 6 rounds of serial transplantation. A total of 200 cells were counted in each sample. The original data are reported in supplemental Table 7. ALL cells with germline, heterozygous deletion, and homozygous deletions are represented by colored dots of size proportional to the percentage of cells in each category in relation to the total cell number as indicated in the legend. One case, VHR-01, shows clonal selection toward additional deletions at this locus, whereas the 4 other cases analyzed are more stable with respect to the clonal composition based on this genetic feature. Original magnifications ×40 (Leica LX Widefield Microscope DMI6000B with the DFC 350 FX R2 camera).
A dominant pattern of Ig/TCR rearrangements is propagated in VHR-ALL xenografts
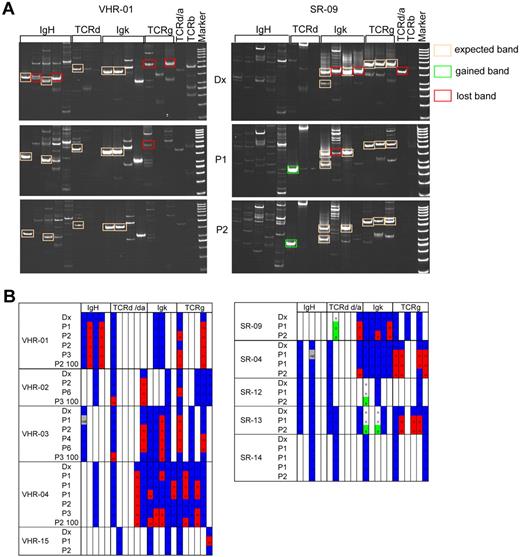
To detect significant changes in the clonal distribution in ALL, we took advantage of the standardized polymerase chain reaction (PCR)-based methodology to detect clonal specific rearrangements of the Ig and TCR genes in ALL xenografts.25 In 10 of 14 patient samples analyzed, composing primary up to quaternary xenografts of VHR- and SR-ALL samples, the 2 markers that were used for MRD follow-up in the patients remained stable in the xenografted samples (supplemental Table 8). In 4 cases, one or both of these MRD markers could not be detected after transplantation. This is reminiscent of a large clinical study that reported the loss of both diagnostic MRD markers at relapse in 11% of patients.38 We then screened 10 cases (5 VHR-ALL and 5 SR-ALL) systematically for the presence of 23 clonal specific Ig/TCR rearrangements in the diagnostic and corresponding xenografts, looking at replica from the same passage number and at serially passaged xenografts (Figure 6). We sequenced all detected bands of the expected size to verify accuracy of the PCR on xenograft material at least once. Two representative examples are shown in Figure 6A, and a summary of all data is represented in a schematic form (Figure 6B; supplemental Figure 2). Overall, the pattern of expected Ig/TCR rearrangements was remarkably stable. In a representative VHR-ALL sample (Figure 6A), 5 markers were stably detected in both the diagnostic and in xenograft samples (boxed in orange in Figure 6A), whereas 4 markers were lost in at least one of the xenograft samples (boxed in red in Figure 6A). In general, a similar pattern of dominant markers could be detected in all xenograft passages, whereas additional markers were lost to a variable degree in xenografts (Figure 6B). Interestingly, in 3 of 5 SR-ALL samples, 1 or 2 new Ig/TCR rearrangements emerged in xenograft samples (Figure 6B). For instance, in SR-9, one new rearrangement was detected as a strong PCR fragment in primary and secondary xenografts, but not in the diagnostic sample (Figure 6A). Interestingly, samples with the appearance of new Ig/TCR rearrangements (SR-9, SR-12, and SR-13) showed the greatest acceleration in engraftment from primary to secondary passage (11 vs 4 weeks, 14 vs 3 weeks, and 28 vs 5 weeks; supplemental Table 2). Whether these rearrangements preexisted in minor subclones at diagnosis or occurred during propagation of the disease in mice cannot be determined based on these data. The Ig/TCR patterns detected in leukemia reconstituted from 100 unsorted cells (VHR01-04) were comparable to the patterns obtained from bulk transplanted samples (Figure 6B). This analysis reinforces the notion that the LIC compartment contains related but distinct clonal subpopulations with a dominant pattern selected in NSG mice.
Analysis of Ig/TCR rearrangements reflects the evolution of the clonal compartment in xenografts. The Ig/TCR rearrangement pattern of diagnostic samples and matched xenografts was determined in 5 VHR- and in 5 SR-ALL samples by PCR using standardized sets of primers that are validated for MRD analysis. (A) Representative examples for a VHR-ALL (left panel) and an SR-ALL (right panel) sample, which showed a marked acceleration from primary to secondary passage. All positive bands of the correct size were sequenced once for verification. Stable rearrangements are framed with an orange line; and bands framed in red represent markers that disappeared in xenografts. In this SR sample, one new Ig/TCR rearrangement appeared (framed in green). (B) Overview of Ig/TCR rearrangements detected. Detected Ig/TCR rearrangements are highlighted in blue. Loss of such a marker in a given xenograft is highlighted in red. Newly detected markers in xenografts are highlighted in green.
Analysis of Ig/TCR rearrangements reflects the evolution of the clonal compartment in xenografts. The Ig/TCR rearrangement pattern of diagnostic samples and matched xenografts was determined in 5 VHR- and in 5 SR-ALL samples by PCR using standardized sets of primers that are validated for MRD analysis. (A) Representative examples for a VHR-ALL (left panel) and an SR-ALL (right panel) sample, which showed a marked acceleration from primary to secondary passage. All positive bands of the correct size were sequenced once for verification. Stable rearrangements are framed with an orange line; and bands framed in red represent markers that disappeared in xenografts. In this SR sample, one new Ig/TCR rearrangement appeared (framed in green). (B) Overview of Ig/TCR rearrangements detected. Detected Ig/TCR rearrangements are highlighted in blue. Loss of such a marker in a given xenograft is highlighted in red. Newly detected markers in xenografts are highlighted in green.
Taken together, our results indicate that the LIC compartment in ALL is large and consists of a mixed population of related subclones. Expansion in NSG mice is associated with clonal evolution to some extent, as there were corresponding subclones present in the diagnostic samples from which xenografts were derived. In many instances, selection appears to occur with the first passage and the patterns of genetic markers remain sufficiently stable to use the xenograft system to model disease.
Discussion
Understanding the nature of the leukemia propagating cell compartment in resistant ALL will greatly impact on the design and experimental evaluation of new treatment rationales. We established a xenograft model with diagnostic samples from patients with highly resistant disease based on in vivo response to treatment, which remains the strongest predictor of poor outcome in ALL.9 Here we provide compelling evidence to propose that both chemoresistant and chemosensitive ALL consist of highly related, albeit genetically diverse, clonal subpopulations. These results corroborate with contemporary studies of ETV6-RUNX1–positive ALL20 and BCR-ABL–positive ALL21 and imply that ALL can be propagated as dynamic multiclonal populations of LICs. Functionally, the analysis of both serial and replica xenografts from several independent diagnostic samples reveals that genetically distinct subclones have different NSG repopulation capacity, suggesting selection of properties conferring selective advantages in NSG mice. In many instances, a pattern reflecting the propagation of highly related clonal populations was detected. Only minimal changes in CNAs occurred, mostly in chromosomal regions that were previously reported in ALL, which indicates that these change are neither the result of increased genetic instability, nor specifically the result of selective pressure from the xenogenic environment. The analysis of clonal Ig/TCR rearrangements revealed loss of less predominant markers, suggesting that most changes may be inferred to the loss of minor ALL subclones.
Some differences emerge between highly resistant VHR-ALL and chemosensitive SR-ALL cases. Most notable is the acceleration of the engraftment kinetics with the second passage into mice, which correlates in 3 of 5 cases with the emergence of readily detectable new Ig/TCR rearrangements. It is tempting to speculate that, in VHR-ALL patients with a very unfavorable leukemia biology, ALL clones have been selected with genetic or epigenetic features that favor engraftment and proliferation in a xenogenic environment. This could explain the propagation of dominant subclones of leukemia cells for VHR-ALL in NSG mice. Similarly, in selected cases of BCR-ABL ALL xenografts, the pattern of genetic lesions was more concordant in cases that engrafted mice readily, which was associated with a trend toward shorter survival times in patients.21 Strikingly, this was strictly associated with additional deletions in the CDKN2A/B locus in BCR-ABL ALL xenografts. In our series, the occurrence of additional deletions at this locus was also observed but occurred both in SR-ALL and VHR-ALL cases. Although genome-wide association studies clearly implicate the CDKN2A/B locus in the pathogenesis of ALL,39 the prognostic value of deletions of this locus in ALL is less clear, as the association with relapse is inconsistent.40 Given that the number of cases included in such xenograft studies is necessarily small and the ALL subgroups compared heterogeneous with respect to their underlying genetic features, we remain cautious with the interpretation of these results. Moreover, the relevant mechanisms to escape the selective pressure of chemotherapy may not necessarily overlap with factors that favor engraftment in a xenogenic environment. Indeed, knowing that in many cases the pattern of clonal-specific genetic markers are dominant over several passage in NSG mice, it will be possible to study the consequences of selective pressure by therapeutic agents or interference with microenvironmental factors with the xenograft system in vivo. As such, this model may be useful to functionally identify driving events that lead to disease progression and resistance to treatment. In particular, the xenograft model may provide the means to evaluate the effect of recurrent mutations that are more frequently detected at relapse in ALL with respect to clonal selection and drug resistance.15
Our data, together with recently published evidence20,21 showing that the leukemogenic compartment is composed of genetically distinct albeit related clones, challenge the hierarchic model, which proposed that the leukemia is maintained from a subset of immature stem cells.16 Our findings that 100 VHR-ALL cells are sufficient to reconstitute leukemia in NSG mice, a strain that is more permissive for lymphoid cells,41 suggests that leukemia propagating cells are not restricted to a rare subset of cells. Similar LIC frequencies were reported for other ALL subtypes.20,21,33 Likewise, the fact that sorted ALL cells reconstitute immunodeficient mice efficiently independent of their immuno-differentiation status5,20 supports this notion. In the absence of markers to enrich for NSG repopulating capacity, definitive conclusions with respect to the hierarchy of LIC in ALL have to be deferred. Our limiting dilution experiments indicate that primary diagnostic samples engraft at a lower frequency than samples that were previously established in NSG. These results may be influenced by differences in cell viability in the stored primary patient samples. However, the trend to accelerated engraftment that we observed with samples from chemosensitive patients on the second passage in NSG suggests that selection for better NSG repopulating capacity occurs in some cases. But the xenograft assay is a reflection of combined functions, which include self-renewal capacity, but also homing and protective interactions with the recipient microenvironment. Further improvements of humanized immunodeficient mouse strains may contribute to clarifying this issue.42 Given 2 other reports that imply an association between NSG repopulation kinetics and adverse outcome,21,43 prospective and detailed investigation is warranted to understand whether leukemia with a more favorable biology is associated with lower frequencies of LIC and to dissect the nature of the selective advantages in HR leukemias. Studies of relapses cases from patients that were classified in the SR group at initial presentation using the xenograft system will be needed to understand whether outgrowth of minor subclones may be clinically relevant in this setting.
Taken together, our results demonstrate that the clonal composition of leukemia samples from patients with highly resistant disease remains remarkably stable on serial passage in NSG mice. Based on available data, it is probable that a larger proportion of ALL cells are leukemogenic. The oligoclonal or even polyclonal nature of ALL may confer the ground for clonal selection/progression to escape chemotherapy. The mouse xenograft system will enable to investigate the consequences of different types of interventions on the clonal composition of the disease. Clearly, development of new therapeutic approaches will have to take into account that the whole leukemia population has to be eradicated for successful treatment. These considerations also reinforce the importance of combination therapy, which is supported by decades of clinical research in oncology. The development of xenograft models of distinct clinical and genetic entities will greatly improve our capability to evaluate new treatment rationales. With this renewable source of relevant samples, combinatorial studies can be performed taking the clonal complexity of the disease into consideration.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Beat Schäfer, Silvia van Essen, and Susanne Kubetzko for instructions and reagents to perform the Ig/TCR experiments and Martina Kähler for performing some of the sequence analysis of Ig/TCR rearrangements.
This work was supported by the Swiss National Science Foundation (grant 310030-118392). J.-P.B. was supported by Claus Cramer Stiftung, Empiris Foundation, Fondation pour la recherche cancer de l'enfant, Hanne-Liebermann Stiftung, Oncosuisse, Novartis Foundation for Biomedical Research, Vontobel-Stiftung, Stiftung zur Krebsbekämpfung, Zurich, and Werner und Hedy Berger-Janser Stiftung zur Erforschung der Krebskrankheiten. This work was also supported in part by the Madeleine Schickedanz-Kinderkrebs-Stiftung and the National Genome Research Net (M. Stanulla and M. Schrappe).
Authorship
Contribution: M. Schmitz, P.B., N.S., L.B., P.M., and B.C.B. performed experiments; M. Stanulla and M. Schrappe provided samples and anonymized clinical information; M. Schmitz contributed to writing the manuscript; B.C.B. and J.-P.B. designed research, analyzed data, and wrote the manuscript; and all authors analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jean-Pierre Bourquin, Division of Oncology and Children's Research Center, University Children's Hospital, University of Zurich, 8032 Zurich, Switzerland; e-mail: jean-pierre.bourquin@kispi.uzh.ch.
References
Author notes
B.C.B. and J.-P.B. contributed equally to this study.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal