Abstract
The IFN-inducible immunity-related p47 GTPase Irgm1 has been linked to Crohn disease as well as susceptibility to tuberculosis. Previously we demonstrated that HSC quiescence and function are aberrant in mice lacking Irgm1. To investigate the molecular basis for these defects, we conducted microarray expression profiling of Irgm1-deficient HSCs. Cell-cycle and IFN-response genes are up-regulated in Irgm1−/− HSCs, consistent with dysregulated IFN signaling. To test the hypothesis that Irgm1 normally down-regulates IFN signaling in HSCs, we generated Irgm1−/−Ifngr1−/− and Irgm1−/−Stat1−/− double-knockout animals. Strikingly, hyperproliferation, self-renewal, and autophagy defects in Irgm1−/− HSCs were normalized in double-knockout animals. These defects were also abolished in Irgm1−/−Irgm3−/− double-knockout animals, indicating that Irgm1 may regulate Irgm3 activity. Furthermore, the number of HSCs was reduced in aged Irgm1−/− animals, suggesting that negative feedback inhibition of IFN signaling by Irgm1 is necessary to prevent hyperproliferation and depletion of the stem cell compartment. Collectively, our results indicate that Irgm1 is a powerful negative regulator of IFN-dependent stimulation in HSCs, with an essential role in preserving HSC number and function. The deleterious effects of excessive IFN signaling may explain how hematologic abnormalities arise in patients with inflammatory conditions.
Introduction
The p47 GTPase Irgm1 is a critical component of murine immunity against a wide variety of pathogenic bacteria and protozoa.1 Irgm1 belongs to the family of immunity-related GTPases (IRGs) that are robustly induced in response to IFN signaling2 and conserved across many species, including zebrafish, mice, and humans.1 In mice the IRG family contains 23 members, of which Irgm1 is arguably the most important because its disruption leads to the broadest susceptibility to bacterial and parasitic infections.1
In contrast to the mouse, humans have only 2 truncated IRG genes, leading some investigators to conclude that these proteins are not used in human immunity.3 However, increasing evidence suggests that IRGM1 is important in human physiology. Knockdown of IRGM1 expression in cultured human macrophages disrupts autophagy-dependent degradation of mycobacterial agents.4 In genome-wide association studies, investigators have shown that mutations in IRGM1 are associated with Crohn disease, an autoimmune condition, and people with mutations in IRGM1 are more susceptible to tuberculosis.5-8
Despite its physiologic importance in the IFN-mediated immune response, the mechanism of Irgm1 action remains poorly understood. Irgm1 is thought to facilitate autophagy-dependent killing of pathogens by the innate immune system, but in protein interactions studies to assess how this is achieved, authors mainly have shown that the murine IRG proteins interact with one another to regulate subcellular localization.9-11 The IRG proteins are associated with endosomal membranes, leading some to speculate that they participate in immune clearance of pathogens by regulating formation of autophagosomes or phagolysosomes.12,13
We previously reported a critical role for Irgm1 in regulating HSC quiescence and function.14 HSCs are pluripotent cells that typically reside in a dormant state in the BM. Under stressful conditions, HSCs can be stimulated to divide and/or differentiate into all of the cell types of the peripheral blood.15 Mice deficient in Irgm1 become anemic and neutropenic as a result of chronic infection, and their HSCs are strikingly defective in the ability to reconstitute the blood of a BM-depleted host.14 We sought to characterize the mechanism of Irgm1-dependent regulation of HSC proliferation and function, with the hope that such knowledge might elucidate the pathophysiology of hematologic abnormalities associated with inflammatory conditions, including autoimmune diseases and chronic infections.
Methods
Mice
Wild-type (WT) C57Bl/6 (CD45.2) and C57Bl/6.SJL (CD45.1) mice 6-12 weeks of age were used. C57Bl/6 Ifngr1−/− (Stock #3288) mice were obtained from The Jackson Laboratory.16,17 C57Bl/6 Stat1−/− mice were obtained from David Levy (New York University).18 GFP-LC3 transgenic mice were obtained from N.T.E. (Baylor College of Medicine).19 C57Bl/6 Irgm1−/−, Irgm3−/−, and Irgm1−/−Irgm3−/− mice were obtained from G.A.T. (Duke University).10,20,21 Irgm1−/− and Stat1−/− strains were intercrossed to produce mice that were homozygous for disruptions at the Irgm1 and Stat1 loci (Irgm1−/−Stat1−/−). Irgm1−/− and Ifngr1−/− strains were similarly intercrossed to produce mice that were homozygous for Irgm1 and Ifngr1 mutations (Irgm1−/−Ifngr1−/−). Irgm1−/− and GFP-LC3 were intercrossed to produce mice that were homozygous for Irgm1 mutation and that expressed the GFP-LC3 transgene (Irgm1−/−LC3+). All mouse strains were maintained at an AALAC-accredited, specific-pathogen-free animal facility at Baylor College of Medicine, and all animal studies were approved by the Baylor College of Medicine Institutional Animal Care and Use Committee.
Immunohistochemistry
HSCs (CD150+ SPKLS) were fixed with 4% paraformaldehyde and permeabilized with 10% Triton-X 100 (Sigma-Aldrich). Cells expressing the GFP-LC3 transgene were directly visualized, whereas cells that did not express the transgene were stained with anti-LC3 antibody (sc-28266; Santa Cruz Biotechnology) and FITC-conjugated anti–rabbit secondary antibodies by standard methods. Cells were visualized with a DeltaVision restoration microscope and images processed with Softworx imaging software.
BM transplantation
Noncompetitive BM transplantations were performed by retro-orbital intravenous injection of 2 × 106 CD45.2 donor whole BM (WBM) cells from 8- to 12-week-old mutant mice or WT controls (C57Bl/6) into 6- to 8-week-old CD45.1 WT C57Bl/6 recipients that had been lethally irradiated with a split dose of 10.5 Gy. For competitive transplants, 2 × 106 CD45.2 donor WBM cells from WT or mutant mice were admixed with 2 × 106 WT CD45.1 competitor WBM cells before they were injected into CD45.1 recipients. Engraftment was measured at 4, 8, 12, and 16 weeks after transplantation by staining of WBCs from peripheral blood with anti-CD45.1 and anti-CD45.2 antibodies.
BrdU labeling
Mice received an intraperitoneal injection of BrdU (Sigma-Aldrich; 1 mg per 6 g of mouse weight), followed by BrdU in drinking water (Sigma-Aldrich; 1 mg/mL) for 3 days. Mice were killed, and CD150+ SPKLS were sorted into carrier B220+ splenocytes, as previously described.14 Samples were analyzed for BrdU incorporation by use of the BrdU Flow Kit (BD PharMingen).
RNA purification and hybridization to microarrays
RNA was isolated from 1.5-6 × 104 HSCs (SP; lineage-negative, Sca1-positive [SPLS]; pooled from several mice) by use of the RNAqueous kit (Ambion), treated with DNAseI, and precipitated with phenol/chloroform/isoamyl alcohol. The RNA was linearly amplified as previously reported.22 In brief, after 2 rounds of T7-based in vitro transcription with the MessageAmp kit (Ambion), RNA was labeled with biotin-conjugated UTP and CTP (Enzo Biotech), fragmented to approximately 50 bp (confirmed by electrophoresis), and hybridized to MOE430.2 chips. Three chips were hybridized per genotype. The raw image and intensity files were generated by the use of GCOS 1.0 software (Affymetrix). All microarrays used in this study had to pass several quality control tests, including scale factor < 5, a 5′ to 3′ β actin probe ratio < 15, % call in MAS 5 (30 < × < 60), and replicate correlation coefficient > 0.96. The arrays well exceeded these standards (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Microarray analysis
Normalization and model-based expression measurements were performed with the gene chip application of robust multiarray average analysis as previously described.22 Control probes were removed, and a representative probe set with the greatest expression magnitude was selected for each gene. The remaining ∼ 11 000 unique genes were used to perform a pair-wise comparison in HSCs with linear models for microarray data. Genes found to have a differential fold-change ≥ 2 and a multiple test correction-adjusted P ≤ .05 were considered to be significantly different between WT and Irgm1−/− HSCs. GC robust multiarray average and linear models for microarray data are available as part of the Bioconductor project (http://www.bioconductor.org) within the programming language R (http://cran.r-project.org/).23 Categorization of differentially expressed genes was accomplished with the Gene Ontology (GO; http://www.geneontology.org) bioinformatics web tool. Gene list enrichment for GO categories was calculated with the FatiGO tool from Babelomics (Babelomics 4.2; http://babelomics.bioinfo.cipf.es/fatigo.html), which calculated significance by the use of the Fisher exact test to determine the false-discovery rate.24
Quantitative real-time RT-PCR
Ccl5 and Ccnb1 Taqman probes were used with Taqman PCR Mastermix and a 7900HT Fast Real-Time PCR system. Samples run in triplicate were normalized to internal 18S controls (Applied Biosystems).
Cytokine detection
IFNγ, CXCL9, and CCL5 levels were detected with the use of a mouse BD cytokine bead array and BD FacsArray plate reader or IFNγ ELISA (BD). BM supernatant was isolated from mouse tibias and femurs by suspending the bones in P200 pipette tips trimmed to fit into 1.5-mL Eppendorf tubes containing 10 μL of PBS and centrifuging them at 500g for 8 minutes. After the discharged marrow was pooled and recentrifuged, the clear supernatant was isolated, and total protein was quantified with a NanoDrop spectrophotometer (NanoDrop Technologies).
Flow cytometry
Generation of constructs and cell lines
Murine Irgm1 was amplified from IMAGE clone 40131260, cloned into the pENTR/D-TOPO vector (Invitrogen), and the Gateway system was used to recombine it into the pBabe-puro retroviral vector. The retroviral vector was cotransfected with pCL-Eco vector into HEK-293 cells to produce retrovirus for the subsequent infection of 32D cells.27 Retroviral transduction was performed as previously described.28 Transduced cells were selected with 2 μg/mL puromycin for 3 days after infection to obtain cells stably expressing full-length Irgm1 tagged with a FLAG epitope at the N terminus. Western blots to confirm stable expression were performed as previously described.29 In brief, protein lysates were isolated from 32D or HEK-293 cells and separated by SDS-PAGE. Irgm1 was detected by Western blot analysis by the use of Irgm1 goat antipeptide antiserum and donkey anti–goat IgG-HRP or anti-FLAG (Sigma-Aldrich) followed by anti–mouse (Calbiochem) secondary antibody.
Pulldown screen for protein-protein interactions
A previously described protocol was used to perform large-scale affinity purification of FLAG-tagged Irgm1.29 In brief, 32D cells with and without stably transduced FLAG-tagged Irgm1 were grown in suspension up to 1 × 106 cells/mL. Protein extracted from this suspension was incubated with M2 soluble anti-FLAG antibody for 2 hours at 4°C. The supernatant was then immunoprecipitated with Protein A/G agarose beads (Santa Cruz Biotechnology) for 1 hour at 4°C. The beads were washed 4 times with NETN (20mM Tris, pH 8.0; 100mM NaCl; 0.5% Nonidet P-40; and 1mM EDTA). The beads were then boiled in SDS loading buffer, separated on a precast 4%-20% SDS-PAGE gradient gel (BioRad), and visualized by Coomassie Blue staining. Bands were excised, digested in trypsin, and subjected to ion trap mass spectrometry as previously described.30 Peptides were identified with PROWL (http://prowl.rockefeller.edu).
Statistics
Mean values ± SEM are shown. Student t-test or ANOVA was used for comparisons (GraphPad Prism Version 5.0); *P < .05, **P < .01, ***P < .001. LC3 aggregation was scored and proportions were compared using a 2-sample test of proportions. One asterisk indicates 90% confidence interval (CI), 2 asterisks 95% CI, and 3 asterisks 99% CI.
Results
Microarray analysis of Irgm1-deficient HSCs reveals increased cell-cycle and IFN-mediated gene expression
We have previously reported that Irgm1-deficient animals become anemic and neutropenic in response to Mycobacterium avium infection, suggesting an HSC defect.31 When we observed the BM of Irgm1−/− mice, we noted that the HSCs were hyperproliferative compared with the HSCs of WT animals.14 This phenomenon is reminiscent of the BM from WT animals that have been exposed to stress conditions such as chemotherapy or infection.22,31 This observation suggests that stimulatory effects on hematopoietic progenitors observed in infected WT animals are already present in naive Irgm1−/− animals.
We also previously noted that the side population of BM cells, a group of cells that efflux Hoechst 33342 dye and that are highly enriched for functionally defined HSCs, was expanded in WT mice in response to M avium infection but that this expansion did not occur in Irgm1−/− mice when they were similarly infected.14 We postulated that the BM of Irgm1-deficient animals lacks responsiveness to M avium infection because HSCs from uninfected Irgm1−/− mice are already influenced by inflammatory signaling.
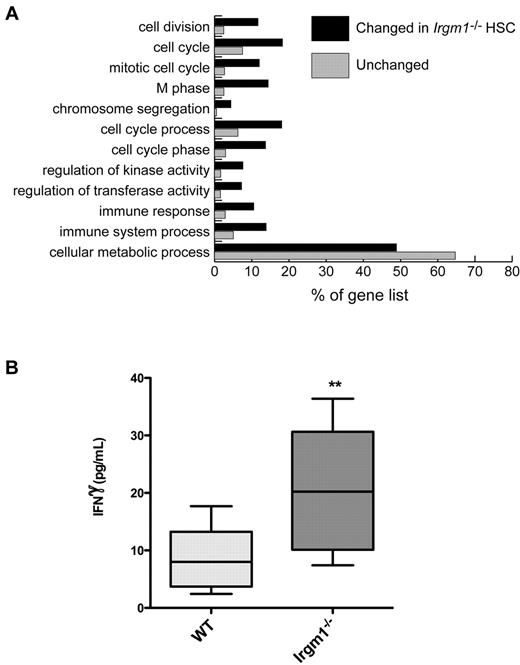
To test this hypothesis, we undertook global gene expression analysis to compare gene transcription between HSCs from WT versus Irgm1-deficient mice. We predicted that inflammatory and cell-cycle pathways would be increased in the Irgm1-deficient cells. We used the FatiGO web tool from Babelomics (Babelomics 4.2) to gain a global perspective of the GO categories differentially expressed by comparing the Irgm1-deficient and WT HSCs. According to this analysis, each gene on the array was designated as “changed” or “unchanged” in the KO versus WT samples. Subsequently, these lists were subdivided among the known GO categories, and the categories are assigned a significance on the basis of the relative abundance of their members in the “changed” as opposed to the “unchanged” groups. This analysis revealed 12 GO categories that met the statistical criteria (P < .05): 11 GO categories were enriched, whereas 1 GO category (“cellular metabolic process”) was underrepresented in Irgm1−/− HSCs compared with WT HSCs (Figure 1A). Of note, 7 of the enriched categories were directly related to cell division and cell cycle, and 2 were related to immune responses (Figure 1A).
The expression profile for Irgm1−/− HSCs shows enrichment of cell proliferation and immune-related GO categories; IFNγ levels are increased. (A) Genes that were differentially expressed in Irgm1−/− HSCs were subjected to an analysis of GO category enrichment by use of the FatiGO webtool (Babelomics 4.2). This tool compared genes differentially regulated in KO HSCs (black bars) against all other genes (unchanged genes: gray bars); these lists were analyzed for assortment into GO categories. Genes from cell-cycle/M-phase and immune categories were overrepresented in the list of differentially expressed genes. Only GO categories that showed a significant difference (P < .05) between the list of genes differentially expressed and those that were not changed in KO HSCs are shown. (B) IFNγ levels in serum from WT and Irgm1-deficient mice were quantified by ELISA. n = 10. Box and whiskers plot represents the 25th-75th percentile and 95% confidence intervals. **P < .01.
The expression profile for Irgm1−/− HSCs shows enrichment of cell proliferation and immune-related GO categories; IFNγ levels are increased. (A) Genes that were differentially expressed in Irgm1−/− HSCs were subjected to an analysis of GO category enrichment by use of the FatiGO webtool (Babelomics 4.2). This tool compared genes differentially regulated in KO HSCs (black bars) against all other genes (unchanged genes: gray bars); these lists were analyzed for assortment into GO categories. Genes from cell-cycle/M-phase and immune categories were overrepresented in the list of differentially expressed genes. Only GO categories that showed a significant difference (P < .05) between the list of genes differentially expressed and those that were not changed in KO HSCs are shown. (B) IFNγ levels in serum from WT and Irgm1-deficient mice were quantified by ELISA. n = 10. Box and whiskers plot represents the 25th-75th percentile and 95% confidence intervals. **P < .01.
We next used an adjusted Fisher test to identify genes that were at least 1.8-fold different (P < .05) from their expression level in WT HSCs, yielding a list of 245 genes that were altered in the absence of Irgm1: 125 up-regulated and 120 down-regulated. (The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE11591 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE11591). We found that genes involved in cell-cycle regulation and in IFN signaling were prominently up-regulated in Irgm1-deficient cells. These include 20 genes known to be up-regulated during cell-cycle progression (eg, Ccnb1, Ccna2) and 17 genes known to be transcriptionally regulated by IFNs (eg, Ccl5, Cxcl10; Table 1). In addition, 2 of the up-regulated genes (Selp and Lilrb4) regulate the production of IFNγ and elaboration of the IFN signaling cascade.33,34 In summary, HSCs lacking Irgm1 display aberrantly increased expression of a subset of cell-cycle and IFN-regulated genes. The most parsimonious mechanistic explanation of these results is that Irgm1 is normally involved in feedback inhibition of IFN signaling in HSCs. Given that IFNs stimulate HSC proliferation,32 we suspect that derepression of IFN signaling may lead to increased expression of cell-cycle progression genes in Irgm1-deficient cells.
Cell-cycle and IFN-regulated genes are up-regulated in Irgm1 HSCs
| Cell-cycle genes . | IFN-associated genes . | ||
|---|---|---|---|
| Gene symbol . | Fold-change . | Gene symbol . | Fold-change . |
| Spag5 | 5.8 | Ccl5 | 30.5 |
| Ncapg | 4.4 | Ifit1 | 17.1 |
| Rrm2 | 4.3 | Cxcl10 | 14.7 |
| Ccnb2 | 4.1 | Lilrb4 | 13.5 |
| Ccna2 | 3.9 | Rsad2 | 12.3 |
| Aurkb | 3.6 | Cxcl9 | 8.5 |
| Nupr1 | 3.4 | Usp18 | 7.9 |
| Nek2 | 3.4 | Oasl1 | 7.6 |
| Kif20a | 3.3 | Ifi27 | 6.9 |
| Tk1 | 3.3 | Cxcl11 | 5.5 |
| Dtl | 3.2 | Ifit3 | 5.1 |
| Ccnb1-rs1 | 3.2 | LOC623121 | 5.1 |
| Cdca3 | 3.1 | Irf7 | 4.6 |
| Ccnb1 | 3.1 | Selp | 4.6 |
| Id3 | 3.0 | Oas2 | 3.5 |
| Gins1 | 3.0 | Ifit2 | 3.4 |
| Ube2c | 3.0 | Fas | 3.4 |
| Stil | 2.9 | Mki67 | 3.3 |
| Id2 | 2.9 | Irga6 | 2.8 |
| Bub1 | 2.8 | ||
| Cell-cycle genes . | IFN-associated genes . | ||
|---|---|---|---|
| Gene symbol . | Fold-change . | Gene symbol . | Fold-change . |
| Spag5 | 5.8 | Ccl5 | 30.5 |
| Ncapg | 4.4 | Ifit1 | 17.1 |
| Rrm2 | 4.3 | Cxcl10 | 14.7 |
| Ccnb2 | 4.1 | Lilrb4 | 13.5 |
| Ccna2 | 3.9 | Rsad2 | 12.3 |
| Aurkb | 3.6 | Cxcl9 | 8.5 |
| Nupr1 | 3.4 | Usp18 | 7.9 |
| Nek2 | 3.4 | Oasl1 | 7.6 |
| Kif20a | 3.3 | Ifi27 | 6.9 |
| Tk1 | 3.3 | Cxcl11 | 5.5 |
| Dtl | 3.2 | Ifit3 | 5.1 |
| Ccnb1-rs1 | 3.2 | LOC623121 | 5.1 |
| Cdca3 | 3.1 | Irf7 | 4.6 |
| Ccnb1 | 3.1 | Selp | 4.6 |
| Id3 | 3.0 | Oas2 | 3.5 |
| Gins1 | 3.0 | Ifit2 | 3.4 |
| Ube2c | 3.0 | Fas | 3.4 |
| Stil | 2.9 | Mki67 | 3.3 |
| Id2 | 2.9 | Irga6 | 2.8 |
| Bub1 | 2.8 | ||
The list of genes differentially expressed in Irgm1−/− HSCs (SPL; filtered on a fold difference of 21.5) in microarrays was manually annotated. Twenty cell-cycle genes and 19 IFN-regulated genes were found to be up-regulated in KO HSCs. These genes are listed here, along with the -fold difference between KO and WT cells.
KO indicates knockout; and WT, wild-type.
To verify the microarray results, we examined protein products from the dysregulated IFN-inducible genes Cxcl9 and Ccl5 in the BM of WT and Irgm1-deficient mice by using cytokine bead arrays. We found that both were significantly increased in Irgm1−/− mice (supplemental Figure 1). When we compared IFNγ protein levels, we found that there was significantly more IFNγ in the serum of Irgm1−/− KO animals compared with WT (Figure 1B). These results suggest that IFNγ production is dysregulated in Irgm1−/− animals and furthermore that the defects in hematopoietic stem cell quiescence and function in Irgm1−/− animals may be attributed to excessive IFNγ.
Irgm1−/− expression defects are rescued in Irgm1−/−Ifngr1−/− and Irgm1−/−Stat1−/− double KO mice
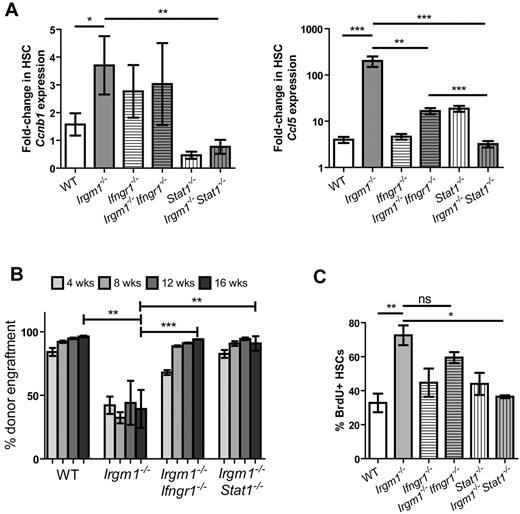
If Irgm1 is involved in negative feedback regulation of IFN signaling, we would expect a double knockout (KO) of Irgm1 and IFN signaling to reverse the phenotypes observed in Irgm1−/− HSCs. Therefore, we generated double mutants lacking both Irgm1 and the IFNγ receptor component Ifngr1 or the major downstream mediator of IFN-induced transcription, Stat1 (Irgm1−/−Ifngr1−/− and Irgm1−/−Stat1−/− double KO mice). We first examined whether the expression of specific genes was normalized in the double KO mice. In the Irgm1 single KO animals, expression of the cell-cycle gene Ccnb1 and the IFN response gene Ccl5 were up-regulated as confirmed by real-time quantitative PCR of cDNA from purified WT or mutant HSCs (Figure 2A). In contrast, we saw a partial rescue of the excessive expression of Ccl5 in HSCs from Irgm1−/−Ifngr1−/− mice and complete rescue of expression of both Ccnb1 and Ccl5 to WT levels in the Irgm1−/−Stat1−/− double KO mice (Figure 2A). These results indicate that aberrantly elevated expression of cell-cycle and chemokine genes in the Irgm1−/− mutant HSCs are manifest only in the presence of intact IFN signaling.
Mutations in Ifngr1 and Stat1 rescue phenotypic and functional defects in Irgm1-deficient HSCs. (A) Gene expression of Ccnb1 and Ccl5 in HSCs (CD150+ SPKLS) from WT, Irgm1−/−, Ifngr1−/−, Irgm1−/−Ifngr1−/−, Stat1−/−, and Irgm1−/−Stat1−/− was analyzed by the use of quantitative RT-PCR. Bars represent n = 3. (B) Noncompetitive BM transplants were performed by the use of WBM from CD45.2 WT, Irgm1−/−, Irgm1−/−Ifngr1−/−, or Irgm1−/−Stat1−/− donors at a dose of 2 × 106 cells. Engraftment was monitored by flow cytometric analysis of peripheral blood chimerism at the indicated time points. n = 3-5 for each bar. (C) HSC proliferation status was assessed by in vivo BrdU labeling. CD150+ SPKLS cells were purified from mice after 3 days of exposure to BrdU. The percentage of CD150+ SPKLS cells that incorporated BrdU during the labeling period is indicated. Bars indicate the mean and SEM and are representative of 2 independent experiments, each performed in triplicate. *P < .05, **P < .01, ***P < .001.
Mutations in Ifngr1 and Stat1 rescue phenotypic and functional defects in Irgm1-deficient HSCs. (A) Gene expression of Ccnb1 and Ccl5 in HSCs (CD150+ SPKLS) from WT, Irgm1−/−, Ifngr1−/−, Irgm1−/−Ifngr1−/−, Stat1−/−, and Irgm1−/−Stat1−/− was analyzed by the use of quantitative RT-PCR. Bars represent n = 3. (B) Noncompetitive BM transplants were performed by the use of WBM from CD45.2 WT, Irgm1−/−, Irgm1−/−Ifngr1−/−, or Irgm1−/−Stat1−/− donors at a dose of 2 × 106 cells. Engraftment was monitored by flow cytometric analysis of peripheral blood chimerism at the indicated time points. n = 3-5 for each bar. (C) HSC proliferation status was assessed by in vivo BrdU labeling. CD150+ SPKLS cells were purified from mice after 3 days of exposure to BrdU. The percentage of CD150+ SPKLS cells that incorporated BrdU during the labeling period is indicated. Bars indicate the mean and SEM and are representative of 2 independent experiments, each performed in triplicate. *P < .05, **P < .01, ***P < .001.
IFN signaling is required for functional defects of Irgm1−/− HSCs
We previously reported that HSCs from Irgm1-deficient animals show an unusually severe impairment in reconstitution assays.14 In fact, these cells fail to reconstitute the BM of lethally irradiated recipients even in the absence of competitor marrow, an experimental condition that is permissive for modest defects in HSC function. To investigate whether this defect is associated with IFN signaling, we tested the performance of HSCs from Irgm1−/−Ifngr1−/− and Irgm1−/−Stat1−/− animals in transplantation assays. In contrast to the severe defects of Irgm1−/− HSCs, HSCs from both double KOs performed as well as WT HSCs in noncompetitive transplants (Figure 2B). In competitive transplantation assays, a more sensitive way of detecting functional differences in the stem cells, Ifngr1 partially rescued the defects of Irgm1 HSCs; however, Stat1 provided complete rescue of this impairment (supplemental Figure 2). These findings indicate that intact IFN signaling is required to confer the functional defect observed in Irgm1-deficient HSCs.
Irgm1−/− phenotypic defects are corrected in Irgm1−/−Ifngr1−/− and Irgm1−/−Stat1−/− double KO mice
To assess whether transcriptional findings in the double mutants also are reflected in phenotypic differences in the cells, we assessed the progenitor compartment (SPL) in single and double KO animals. As previously mentioned, Irgm1-deficient single KO animals display a densely populated SP that is shifted into the upper portion of the distribution, similar to what is seen in WT animals infected with M avium.32 In contrast, there is a partial rescue of the total SPL compartment percentage in the Irgm1−/−Ifngr1−/− animals and full rescue in Irgm1−/−Stat1−/− double KO animals (supplemental Figure 3). There was no expansion in the number of hematopoietic progenitors (SPL) in mice lacking either Ifngr1 or Stat1 when the mice were infected with M avium, an expected result because intact IFN signaling is required for this expansion phenotype (supplemental Figure 4). The finding that the upward-shifted distribution of cells in the SPL compartment of Irgm1 is rescued in the double KO animals is again consistent with a requirement for intact IFN signaling in the phenotype of Irgm1-deficient cells.
In previous work we demonstrated that BM progenitors (Sca1+ SP) are more highly proliferative in Irgm1-deficient mice compared with WT.14 We confirmed our earlier result by using a highly purified subset of HSCs (CD150+ SPKLS)25 from WT and Irgm1-deficient mice that had been treated with BrdU for 3 days. The percentage of cells containing BrdU was substantially increased (78.1% vs 23.0%) in the Irgm1-deficient mice compared with WT, confirming that LT-HSCs from Irgm1-deficient animals are more proliferative than those of WT mice (Figure 2C). When a similar analysis was completed with the use of Irgm1−/−Ifngr1−/− animals, we found partial rescue of the Irgm1 phenotype. In Irgm1−/−Stat1−/− double KO animals, the phenotype was more fully rescued (Figure 2C; supplemental Figure 5). These results were consistent with the gene expression and transplantation results, indicating that abnormalities in Irgm1-deficient HSCs are due to dysregulated IFN signaling.
Irgm1 regulates IFN-induced macroautophagy in the HSC
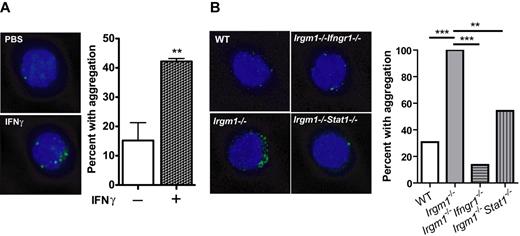
The authors of previous studies have shown that Irgm1 protects CD4+ T lymphocytes from IFNγ-induced macroautophagy and cell death.14 Thus, we hypothesized that Irgm1 may regulate IFNγ signaling in the HSC through modulation of autophagy flux. To study autophagy in HSCs, we examined cytoplasmic aggregation of LC3, a canonical marker for autophagy.35 We found that IFNγ tends to induce LC3-GFP to form cytoplasmic aggregates in HSCs (CD150+ SPKLS) from transgenic LC3-GFP mice,19 which is consistent with alteration of autophagy flux, either through induction of autophagosomes or reduction in their breakdown (Figure 3A). Consistent with high IFNγ levels in animals infected with M avium, we also found that LC3 aggregates were induced in HSCs of M avium-infected animals (supplemental Figure 6).
Irgm1-deficient HSCs exhibit excessive IFN-induced autophagy. (A) LC3-GFP HSCs (CD150+ SPKLS) were isolated and treated with IFNγ in vitro. Representative examples as well as proportions of cells with aggregation are shown for all experiments, each repeated 2 or 3 times with at least 40 cells visualized per cohort. (B) HSCs (CD150+ SPKLS) were isolated from WT, Irgm1−/−, Irgm1−/−Ifngr1−/−, and Irgm1−/−Stat1−/− mice and analyzed for LC3 aggregation by immunofluorescence. Data are representative of 2 independent experiments. *Indicates that the proportions are statistically different with 95% confidence according to a 2-sample test of proportions with P < .05.
Irgm1-deficient HSCs exhibit excessive IFN-induced autophagy. (A) LC3-GFP HSCs (CD150+ SPKLS) were isolated and treated with IFNγ in vitro. Representative examples as well as proportions of cells with aggregation are shown for all experiments, each repeated 2 or 3 times with at least 40 cells visualized per cohort. (B) HSCs (CD150+ SPKLS) were isolated from WT, Irgm1−/−, Irgm1−/−Ifngr1−/−, and Irgm1−/−Stat1−/− mice and analyzed for LC3 aggregation by immunofluorescence. Data are representative of 2 independent experiments. *Indicates that the proportions are statistically different with 95% confidence according to a 2-sample test of proportions with P < .05.
Because Irgm1-deficient mice exhibit excessive IFN signaling at baseline, we anticipated finding dysregulated autophagy in the KO HSCs. We therefore crossed LC3-GFP transgenic mice with Irgm1−/− mice and analyzed their HSCs for LC3 aggregation. We found increased aggregation in HSCs from Irgm1−/− KO transgenic LC3-GFP animals (Figure 3B). In keeping with the abrogation of other phenotypes associated with Irgm1 mutation by mutations in Ifngr1 and Stat1, HSCs from both double KOs exhibited LC3 aggregation levels comparable with WT HSCs as determined by immunohistochemical analysis (Figure 3B). Again, these results indicate that intact IFN signaling is required for the phenotypic defect of increased LC3 aggregation, associated with dysregulated autophagy, seen in Irgm1−/− HSCs.
Interaction between Irgm1 and Irgm3 is required for Irgm1 activity in HSCs
We speculated that identification of Irgm1 binding partners under conditions of IFNγ activation might further elucidate the function of Irgm1 in the HSC. Therefore, we performed large-scale affinity purification of FLAG-tagged Irgm1 via immunoprecipitation of cell lysate and ion trap mass spectrometry as previously described (supplemental Figure 7).30 Peptides were identified by the use of the protein sequence search tool PROWL (supplemental Table 2). Among the 78 proteins that were uniquely pulled down from Irgm1-FLAG lysates, we identified the Irgm1 family member Irgm3, consistent with both yeast-2-hybrid and genetic data from other studies.10,11
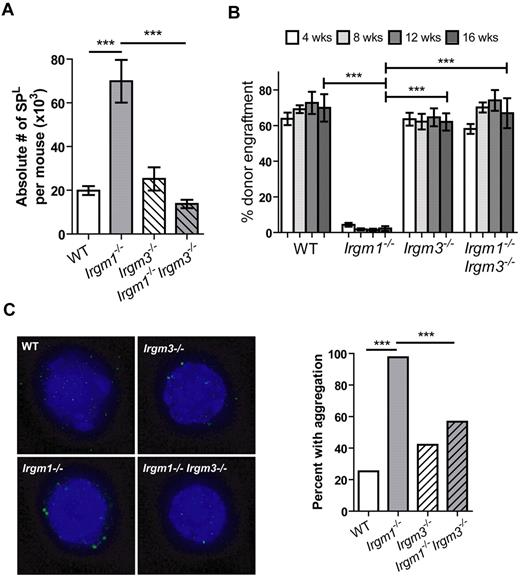
On the basis of these findings, we anticipated that Irgm1 and Irgm3 might interact genetically to regulate HSC function. We performed flow cytometry analysis of WT, Irgm1−/−, Irgm3−/−, and Irgm1−/−Irgm3−/− BM and found that knocking-out both Irgm1 and Irgm3 corrects the phenotype of increased hematopoietic progenitors (expanded SPL) seen in Irgm1 KOs (Figure 4A; supplemental Figure 8A). These findings are similar to what we saw in double KOs for Irgm1 and IFNγ signaling mutants. In addition, we performed WBM transplantation and found that the functional impairment found in Irgm1−/− HSCs is fully normalized in Irgm1−/−Irgm3−/− HSCs in both noncompetitive and competitive transplantation assays (Figure 4B; supplemental Figure 8B). Finally, we assessed LC3 aggregation in HSCs of single and double KO mice. Whereas LC3 aggregation was increased in Irgm1−/− HSCs, this phenomenon was rescued in Irgm1−/−Irgm3−/− HSCs (Figure 4C). Thus, Irgm3 deficiency completely rescues the phenotypic defects seen in Irgm1 HSCs, suggesting that Irgm1 and Irgm3 have opposite or competing functions in HSCs and may negatively regulate each other.
Irgm1−/− HSC function is rescued by the addition of homozygous disruption of Irgm3. (A) The absolute number of SPL cells per mouse is indicated. Data are representative of 2 experiments, each performed in triplicate. (B) Competitive BM were performed with the use of 2 × 106 WBM cells from CD45.2 WT, Irgm1−/−, Irgm3−/−, or Irgm1−/−Irgm3−/− donors admixed with 2 × 106 WBM competitor cells from CD45.1 WT mice. Data are representative of 2 independent experiments, each with 5-6 recipients per cohort. (C) LC3 aggregation was determined by immunohistochemistry in HSCs (CD150+ SPKLS) of WT, Irgm1−/−, Irgm3−/−, or Irgm1−/−Irgm3−/− mice. A minimum of 40 cells was assessed per genotype, and the percentage of cells with LC3 aggregation is shown in the bar graph. Data are representative of 2 independent experiments. The proportions are statistically different with 95% confidence according to a 2-sample test of proportions. ***P < .001.
Irgm1−/− HSC function is rescued by the addition of homozygous disruption of Irgm3. (A) The absolute number of SPL cells per mouse is indicated. Data are representative of 2 experiments, each performed in triplicate. (B) Competitive BM were performed with the use of 2 × 106 WBM cells from CD45.2 WT, Irgm1−/−, Irgm3−/−, or Irgm1−/−Irgm3−/− donors admixed with 2 × 106 WBM competitor cells from CD45.1 WT mice. Data are representative of 2 independent experiments, each with 5-6 recipients per cohort. (C) LC3 aggregation was determined by immunohistochemistry in HSCs (CD150+ SPKLS) of WT, Irgm1−/−, Irgm3−/−, or Irgm1−/−Irgm3−/− mice. A minimum of 40 cells was assessed per genotype, and the percentage of cells with LC3 aggregation is shown in the bar graph. Data are representative of 2 independent experiments. The proportions are statistically different with 95% confidence according to a 2-sample test of proportions. ***P < .001.
Dysregulation of IFN signaling leads to decreased number of HSCs with age
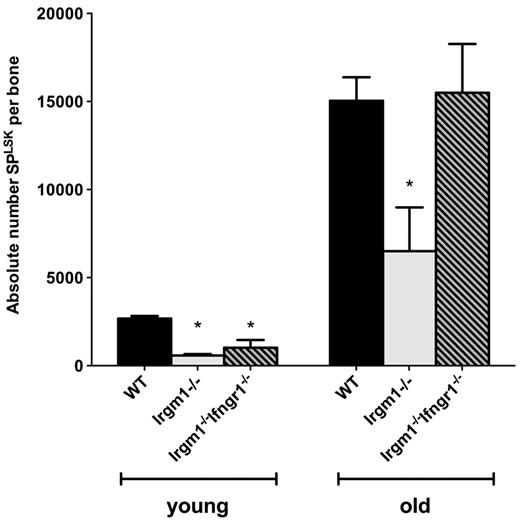
Throughout life, the HSC compartment must be maintained through a careful balance between self-renewal and differentiation events. Imbalance of this system could lead to an insufficient number of HSCs over the long term. Because IFN signaling is known to activate HSC proliferation,32 we postulated that chronically dysregulated IFN signaling, such as that seen in Irgm1−/− mice, might lead to diminished self-renewal and, ultimately, depletion of the HSC compartment. To investigate this hypothesis, we examined the BM of young versus aged WT and Irgm1−/− mice and characterized their HSC subsets. On analysis of aged 12-month-old Irgm1−/− mice, we found that the numbers of HSCs (SPKLS) in aged mice were depleted compared with those of age-matched WT controls, with an average reduction in HSC number by 57% (P < .05; Figure 5). Furthermore, the depletion in HSC number was not seen in 12-month-old Irgm1−/−Ifngr1−/− double KO mice. These findings suggest that dysregulated IFN signaling can lead to depletion of the HSC compartment over time.
The absolute number of HSCs is depleted in the BM of aged Irgm1−/− mice. The absolute number of HSCs (SPLSK) per bone is indicated. Mice were either 8-10 weeks (young) or 12 months of age (old) for all genotypes. For each bar, n = 3. Data are representative of 2 independent experiments. *P < .05.
The absolute number of HSCs is depleted in the BM of aged Irgm1−/− mice. The absolute number of HSCs (SPLSK) per bone is indicated. Mice were either 8-10 weeks (young) or 12 months of age (old) for all genotypes. For each bar, n = 3. Data are representative of 2 independent experiments. *P < .05.
Discussion
Role of Irgm1 highlights the critical nature of IFNγ in regulating HSC activity
The authors of recent studies have demonstrated that IFN signaling can be a powerful activator of HSC proliferation, either in the context of infection or polyinosinic:polycytidylic acid (pIpC) treatment.26,32 Indeed, exposure to IFNγ alone has been shown to be sufficient for activating HSC proliferation, accompanied by functional impairment of the stem cells. In addition, we previously reported that the IFN-regulated GTPase Irgm1 is up-regulated in HSCs during recovery from chemotherapeutic stress and that deletion of Irgm1 leads to extreme functional impairment of the cells. However, the relationship between Irgm1 and IFN-mediated activation of HSCs was not understood.
Here, we have shown that defects in Irgm1-deficient HSCs are because of dysregulation of IFNγ signaling. Thus, Irgm1 provides a critical negative feedback mechanism for IFN production and signaling in HSCs. Absence of this feedback inhibition leads to deleterious effects on HSC function, with diminished ability to expand in response to infection and severely impaired function in BM reconstitution assays. These defects ultimately lead to a depletion of the absolute number of HSCs over time. Furthermore, we have previously found that inflammatory conditions such as chronic infection can amplify these effects, leading to premature BM failure and death in Irgm1-deficient animals.14 These findings are consistent with mouse models of BM failure syndromes in which high IFNγ levels induced by lymphocyte infusion lead to BM hypocellularity and pancytopenia.37 Collectively, these findings highlight the power of the IFN-mediated activation response in HSCs and underscore the importance of careful regulation of IFN signaling in these cells.
We postulate that IFNs released during infection are an important stimulus for HSC activation for the purposes of replenishing the supply of immune cells in the short term. However, this activation response must be suppressed subsequently to avoid deleterious effects of prolonged and unopposed IFN stimulation in the long run. In this study, we have demonstrated that Irgm1 can fulfill this regulatory role in mice. Aside from Irgm1, there are likely to be other mechanisms of inhibitory feedback on IFN signaling in hematopoietic cells. For example, Adar1 similarly represses global IFN responses and is essential for murine hematopoiesis at both embryonic and adult stages.38
Interestingly, we have shown that a mutation in Ifngr1 leads to partial rescue of the Irgm1 phenotypes, including a partial return to WT HSC gene expression patterns, proliferation, and engraftment in transplant assays. In contrast, a Stat1 mutation fully rescues the Irgm1 KO phenotypes (supplemental Figure 2). The Stat1 transcription factor is used for both IFNγ and IFNα/β signaling pathways. It has previously been shown that both IFNγ and IFNα/β can activate dormant HSCs.31,35 Our results suggest that Irgm1-deficient HSCs exhibit dysregulated signaling through both the IFNγ and IFNα signaling pathways; therefore, the Ifngr1 mutation partially rescues the Irgm1-deficient phenotype, whereas the Stat1 mutation leads to a complete rescue likely through interruption of both IFNγ and IFNα signaling.
Our investigation of the role of Irgm1 in HSC biology led us to observe that IFN-inducible autophagy can be activated in HSCs. IFNs have been previously shown to induce autophagy in downstream hematopoietic cells such as macrophages and mature lymphocytes,9,14 but our study represents the first demonstration of autophagy induction in HSCs. Irgm1 was previously found to protect CD4+ T lymphocytes from IFNγ-induced autophagy.14 Our study suggests that there is a relationship between autophagy and HSC function. One possibility is that IFN levels themselves are regulated by autophagic degradation, perhaps through regulation of IFN regulatory proteins Selp and Lilrb4. Hence, in the absence of appropriate autophagy flux IFNγ may accumulate in the serum because of overabundance of these factors. Indeed, a recent study finds that autophagy is important in the maintenance of HSC populations.39 Further studies are needed to delineate the molecular regulation of this process in HSCs.
We have also provided additional evidence for a physical and genetic interaction between Irgm1 and another IRG family member, Irgm3. Functional and phenotypic characteristics of Irgm1-deficient HSCs were rescued in Irgm1−/−Irgm3−/− double KO cells. Irgm3 has previously been implicated in autophagy regulation; macrophages from Irgm3-deficient mice show impaired envelopment of parasites in autophagosome-like vacuoles.40 We speculate that autophagy may function to help HSCs survive by adapting to external stress, rather than to act in direct pathogen killing as is seen in macrophages. Thus, the IRG proteins may serve collectively to assist the host immune response at a systemic level that extends beyond direct pathogen killing.
Although our studies have focused on the murine Irgm1, we anticipate these results also have implications in humans. Several human diseases affecting BM function have been linked to excessive inflammatory signaling. For instance, acquired aplastic anemia syndromes have been linked to conditions of excessive IFNγ release.41 In addition, BM suppression is a known complication of mycobacterial diseases such as tuberculosis, in which IFNγ signaling is high.42 Given our findings in Irgm1−/− mice, we speculate that the tight control of IFN signaling is as important for human HSCs as it is in the murine system. Whether the human homolog of Irgm1, IRGM, fulfills this function remains controversial. Human IRGM has been implicated in autophagy as well as in the inflammatory autoimmune disorder Crohn disease, findings that suggest that it could play a role in regulation of inflammation.4,7 However, the human protein has not been shown to be IFN responsive as is its murine counterpart.3 Future studies will be necessary to understand the regulation of IFN responses in the human hematopoietic system and to determine whether these pathways may be considered as therapeutic targets for patients with immune-mediated BM failure or those with extensive IFN exposure such as during chronic infections.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank David Levy at New York University for providing us with Stat1 KO mice. They are grateful to Yayun Zheng for technical assistance and to the Dan L. Duncan Cancer Center, the Integrated Microscopy Core, the Cytometry and Cell Sorting Core, and the Cancer Center Pathway Discovery Core of Baylor College of Medicine.
K.Y.K. was supported by the Simmons Foundation Collaborative Research Fund and the NHLBI of the National Institutes of Health, and M.T.B. was supported by the NIDDK institute of the National Institutes of Health. Z.S. is a Leukemia & Lymphoma Society Scholar. This work was supported by National Institutes of Health grant AI57831 and a VA Merit Review grant to G.A.T., NIGMS GM081627 and a Welch Foundation Q-1673 grant to Z.S., and by grants of NIDDK, NHLBI, and NIBIB (HL081007, EB005173, DK58192, P50CA126752) to M.A.G.
National Institutes of Health
Authorship
Contribution: K.Y.K. and M.T.B. performed all flow cytometry and cytokine analyses, constructed double mutant strains, analyzed the data, and wrote the paper; K.Y.K. performed immunohistochemistry and microscopy; M.T.B. and G.L.L. performed pull-down assays and coordinated protein chemistry experiments; D.C.W., S.M.C., and N.C.B. performed microarray experiments and analyzed the microarray data; S.W. assisted with real-time experiments; G.L.L. performed protein expression and Western blots; S.Y.J., J.Q., D.L., and Z.S. performed mass spectrometry protein–protein experiments and analysis; N.T.E. provided LC3-GFP mice and assisted with design of autophagy experiments; G.A.T. generated Irgm1−/−Irgm3−/− DKO mice and provided marrow for Irgm3-experiments; and M.A.G. oversaw all experimental design and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for S.M.C. is Sloan-Kettering Institute, New York, NY. The current affiliation for N.C.B. is New York State University, Albany, NY.
Correspondence: Margaret A. Goodell, One Baylor Plaza, N1030, Houston, TX 77030; e-mail: goodell@bcm.edu.
References
Author notes
K.Y.K. and M.T.B. contributed equally to this work.